
Locked Nucleic Acid Oligonucleotide
Properties and application
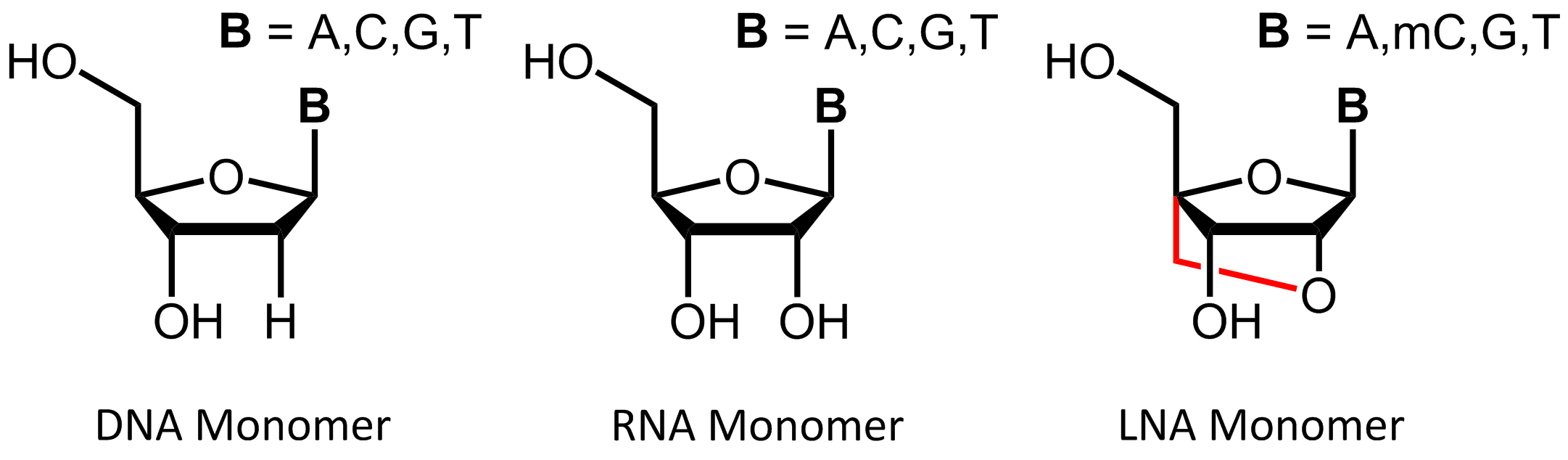
Locked Nucleic Acid (LNA)
Locked Nucleic Acids (LNA) were first independently described in 1997 by Jesper Wengel1 and Takeshi Imanishi2 and have become an important part of hybridisation-based applications.
LNAs are a class of modified RNA-nucleic acid analogues that have an additional methylene bridge. This linkage connects the oxygen atom at the C-2´ of the ribose with the 4´-carbon atom and helps to fix the ribose ring in the
3'-endo conformation ("locked") which leads to the characteristic A-RNA structure and to the exclusive formation of
A-duplexes.
Hybridisation properties and stability
In addition to the common bases adenine, guanine and thymine, methylcytosine is also used in the synthesis of LNA oligonucleotides, allowing hybridisation with complementary DNA or RNA base pairs. The rigid structure of the LNA due to the additional methylene bridge leads to a conformational restricted form that represents the ideal confirmation for Watson-Crick binding. According to this, LNA-based oligos can bind faster to a complementary strand and can also highly increase the stability of the resulting duplex.3
Another notable feature of LNA is its increased affinity for complementary DNA and RNA, which makes LNA-based oligonucleotides ideal for the detection of short and very similar DNA and RNA targets.
Furthermore, the incorporation of individual LNA bases leads to an increase in the melting temperature.3,6 Thus, in experiments, duplexes of LNA:DNA hybrids showed higher annealing temperatures compared to corresponding DNA:DNA duplexes.
Additionally, the incorporation of LNA can increase endo- and exonuclease resistance, allowing high in vivo and in vitro stability of the oligos.5 For example, three LNA bases at both ends of the oligo can protect the oligonucleotide from nucleolytic degradation.7 Probes composing of alternating DNA and LNA building blocks (starting with a LNA base at the 5´-end) show greatly enhanced stability in the cell (nuclease resistance).8
The biostability of LNA-based oligonucleotides in blood and other biological fluids makes them to an interesting candidate for in vivo experiments.6
Specificity and sensitivity
The fact that LNA bases are linked by the same phosphate backbone as native DNA or RNA allows for the synthesis of mixed oligonucleotides consisting of LNA/DNA or LNA/RNA bases. LNA oligonucleotides are synthesised using conventional phosphoamidite chemistry, enabling an automated synthesis of both pure LNA and mixed LNA/DNA or LNA/RNA oligonucleotides.4 In oligonucleotides, one or more LNAs can be incorporated with DNA or RNA nucleotides, thus improving the specificity of the oligos.
The incorporation of a single LNA nucleotide can thereby significantly increase the melting temperature of the oligo, so that, depending on the application, a more precise adjustment of the desired melting temperature (e.g. primer or probe) can be achieved.
Increase of melting temperature Tm
For each incorporation of LNA nucleotides into a DNA oligonucleotide (primer, qPCR probe, etc.) the Tm increases by 2-6°C, into an RNA primer by 3-9.6°C.12 In this way, even short LNA oligonucleotides reach a comparatively high melting temperature and are therefore ideal for the detection of short or very similar target sequences.
Also, AT-rich oligonucleotides showing low melting temperatures can increase their Tm by incorporation of LNA nucleotides.
Tm Normalisation
For some applications where binding to many different targets occurs simultaneously (e.g. microarrays), a narrow Tm range is beneficial. However, differences in the GC content of the target sequences (e.g. miRNA) often complicate detection and analysis with traditional detection methods.
However, the targeted incorporation of LNA nucleotides can adjust the Tm of all probes (Tm normalisation) so that specific binding of different targets under the same reaction conditions is possible.
Further advantages in the application
Comparative studies have shown that LNA/DNA hybrids, unlike PMOs (phosphorodiamidate morpholino oligo, neutral charged backbone), more easily enter cells due to their negatively charged backbone and thereby also allow intracellular applications, e.g. inhibition of translation using LNA antisense oligonucleotides (siLNA).5
An important practical advantage of using LNA oligos is that they are soluble in water, which greatly simplifies their handling.4
Where do LNA oligonucleotides apply?
Because of their diverse properties, LNA oligonucleotides are used for a variety of applications:
- PCR / qPCR applications
- Dual-labelled Probes, Molecular Beacons, Scorpions, also multiplexing is possible
- Allele-specific PCR - in situ Hybridisation (LNA-FISH)
- Microarrays
- Detection of microRNA sequences
- improved differentiation of different microRNA families (regardless of GC content) - Detection of single nucleotide mismatches (SNP detection)
- Gene expression analysis
- Antisense oligonucleotides (gene silencing with siLNA)
- Aptamers
- Pathogen detection
- Methylation analysis
- Mutagenesis studies
Literature:
1. LNA (Locked Nucleic Acids): Synthesis of the adenine, cytosine, guanine, 5-methylcytosine, thymine and uracil bicyclonucleoside monomers, oligomerisation, and unprecedented nucleic acid recognition. Koshkin AA, Singh SK, Nielsen P, Rajwanshi VK, Kumar R, Meldgaard M, Olsen CE, Wengel J; Biochemistry (2006), 45 (23), S. 7447–7455; doi:10.1021/bi060307w.
2. Synthesis of 2′-O,4′-C-methyleneuridine and -cytidine. Novel bicyclic nucleosides having a fixed C3, -endo sugar puckering. Obika S, Nanbu D, Hari Y, Morio KI, In Y, Ishida T, Imanishi T; Tetrahedron Letters (1997), 38 (50), S. 8735–8738; doi:10.1016/S0040-4039(97)10322-7.
3. Structures, dynamics, and stabilities of fully modified locked nucleic acid (β-D-LNA and α-L-LNA) duplexes in comparison to pure DNA and RNA duplexes. Suresh G, Priyakumar UD; J Phys Chem B. (2013), 117(18):5556-64. doi: 10.1021/jp4016068.
4. FISHing with locked nucleic acids (LNA): evaluation of different LNA/DNA mixmers. Silahtaroglua AN, Tommerupa N, Vissing H; Molecular and Cellular Probes 17 (2003) 165–169.
5. LNA/DNA mixmer-based antisense oligonucleotides correct alternative splicing of the SMN2 gene and restore SMN protein expression in type 1 SMA fibroblasts. Touznik A, Maruyama R, Hosoki K, Echigoya Y, Yokota T; Sci Rep. (2017); 7: 3672, doi: 10.1038/s41598-017-03850-2.
6. Biological Activity and Biotechnological Aspects of Locked Nucleid Acids. Lundin KE, Højland T, Hansen BR, Persson R, Bramsen JB, Kjems J, Koch T, Wengel J, Smith CI; Adv Genet. (2013), 82:47-107. doi: 10.1016/B978-0-12-407676-1.00002-0.
7. Design of antisense oligonucleotides stabilized by locked nucleic acids. Kurreck J, Wyszko E, Gillen C, Erdmann VA; Nucleic Acids Res. (2002); 30(9): 1911–1918.
8. Detection of mRNA in living cells by double-stranded locked nucleic acid probes. Riahi R, Dean Z, Wu TH, Teitell MA, Chiou PY, Zhang DD, Wong PK; Analyst. (2013), 138(17):4777-85. doi: 10.1039/c3an00722g.
9. LNA (Locked Nucleic Acid): High-Affinity Targeting of Complementary RNA and DNA. Vester B, Wengel J; Biochemistry (2004), 43 (42), pp 13233–13241, DOI: 10.1021/bi0485732.
10. Locked nucleic acid (LNA): fine-tuning the recognition of DNA and RNA. Braasch DA, Corey DR; Chemistry & Biology (2001), Vol. 8, pp 1-7.
11. Enhanced allele‐specific PCR discrimination in SNP genotyping using 3′ locked nucleic acid (LNA) primers. Latorra D, Campbell K, Wolter A, Hurley JM; Human Mutation (2003), Volume 22, pp 79-85.
12. Oligonucleotide-based systems: DNA, microRNAs, DNA/RNA aptamers. Jolly P, Estrela P, Ladomery M; Essays Biochem. (2016), 60(1): 27–35, doi: 10.1042/EBC20150004.
Design
General Rules for the Design of Locked Nucleic Acids (LNA)
In contrast to standard DNA or RNA oligonucleotides, oligonucleotides with LNA bases tend to be shorter due to their significantly higher melting temperature and the associated higher sensitivity.
When designing oligonucleotides with LNA bases, the general rules for the design of "normal" oligonucleotides apply initially:
- The GC content should be between 30-60%.
- For primer pairs, the values of the Tm should be nearly equal.
In addition, the following aspects can be considered as rough guidelines:
- More than 4-5 consecutive LNA bases (except of very short oligos) and 3 +G or +C stretches should be avoided.1
- At the 3´-end or near the 3´-terminus, several LNA bases should not be positioned.1
- LNA bases should not be within palindromic sequences, since in this case the binding within the oligos may be stronger than to the actual target.2
- Upon binding to DNA, the incorporation of a single LNA building block can increase the Tm by 2-6°C and by 3-9.6°C upon binding to RNA.5
- If several LNA building blocks are used per oligonucleotide, the influence of an LNA monomer on the Tm decreases.
- Terminal LNA bases have only little effect on Tm.3
- The Tm is not or hardly affected if the oligonucleotide has only one LNA building block at the 5´-end.4
- Overall, more than 15 LNA bases within an oligos should be avoided as this promotes self-hybridisation. For longer oligonucleotides (> 15 bases), the LNA content is correspondingly reduced.
Literature:
General information
1. Biological Activity and Biotechnological Aspects of Locked Nucleid Acids. Lundin KE, Højland T, Hansen BR, Persson R, Bramsen JB, Kjems J, Koch T, Wengel J, Smith CI; Adv Genet. (2013), 82:47-107. doi: 10.1016/B978-0-12-407676-1.00002-0.
2. Single nucleotide polymorphism genotyping using short, fluorescently labeled locked nucleic acid (LNA) probes and fluorescence polarization detection. Simeonov A, Nikiforov TT; Nucleic Acids Res. (2002), 30(17):e91.
3. Design of antisense oligonucleotides stabilized by locked nucleic acids. Kurreck J, Wyszko E, Gillen C, Erdmann VA. Nucleic Acids Res. 2002 May 1;30(9):1911-8.
4. Stopped-flow kinetics of locked nucleic acid (LNA)-oligonucleotide duplex formation: studies for LNA-DNA and DNA-DNA interactions. Christensen U, Jacobsen N, Rajwanshi VK, Wengel J, Koch T; Biochem J. (2001); 354(Pt 3):481-4.
5. Oligonucleotide-based systems: DNA, microRNAs, DNA/RNA aptamers. Jolly P, Estrela P, Ladomery M; Essays Biochem. (2016), 60(1):27-35. doi: 10.1042/EBC20150004.
6. Comparison of different real-time PCR chemistries and their suitability for detection and quantification of genetically modified organisms. Buh Gasparic M, Cankar K, Zel J, Gruden K; BMC Biotechnol. (2008), 8:26. doi: 10.1186/1472-6750-8-26.
7. Novel locked nucleic acid (LNA)-based probe for the rapid identification of Chlamydia suis using real-time PCR. Lis P, Kumala A, Spalinski M, Rypula K; BMC Vet Res. (2014), 10: 225. doi: 10.1186/s12917-014-0225-4.
in situ Hybridisation
8. Detection of mRNA in living cells by double-stranded locked nucleic acid probes. Riahi R, Dean Z, Wu TH, Teitell MA, Chiou PY, Zhang DD, Wong PK; Analyst. (2013), 138(17):4777-85. doi: 10.1039/c3an00722g.
9. In situ detection of miRNAs in animal embryos using LNA-modified oligonucleotide probes. Kloosterman WP, Wienholds E, de Bruijn E, Kauppinen S, Plasterk RH; Nat Methods. (2006), 3(1):27-9.
10. A Sensitive Alternative for MicroRNA In Situ Hybridizations Using Probes of 2′-O-Methyl RNA + LNA. Søe MJ, Møller T, Dufva M, Holmstrøm K; J Histochem Cytochem. (2011), 59(7): 661–672, doi: 10.1369/0022155411409411.
Antisense oligos
11. Potent and nontoxic antisense oligonucleotides containing locked nucleic acids. Wahlestedt C, Salmi P, Good L, Kela J, Johnsson T, Hökfelt T, Broberger C, Porreca F, Lai J, Ren K, Ossipov M, Koshkin A, Jakobsen N, Skouv J, Oerum H, Jacobsen MH, Wengel J; Proc Natl Acad Sci U S A. (2000), 97(10):5633-8.
SNP Microarray Analysis
12. Design of LNA probes that improve mismatch discrimination. You Y, Moreira BG, Behlke MA, Owczarzy R; Nucleic Acids Res. (2006), 34(8):e60.
Molecular Beacons
13. Synthesis and investigation of deoxyribonucleic acid/locked nucleic acid chimeric molecular beacons. Yang CJ, Wang L, Wu Y, Kim Y, Medley CD, Lin H, Tan W; Nucleic Acids Res. (2007), 35(12): 4030–4041, doi: 10.1093/nar/gkm358.
Hydrolysis probes
14. A locked nucleic acid (LNA)-based real-time PCR assay for the rapid detection of multiple bacterial antibiotic resistance genes directly from positive blood culture. Zhu L, Shen D, Zhou Q, Li Z, Fang X, Li QZ; PLoS One. (2015), 10(3):e0120464. doi: 10.1371/journal.pone.0120464.
Ordering information
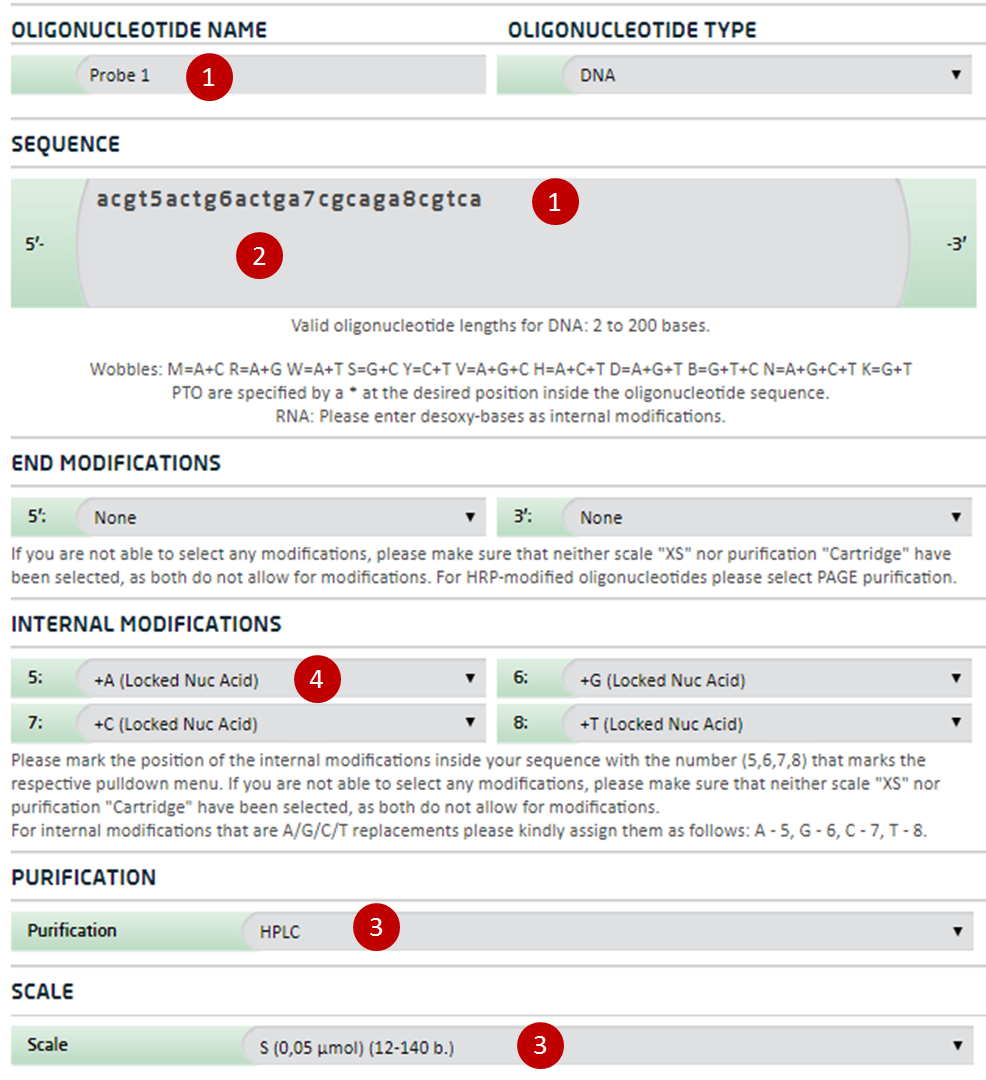
Ordering information for Locked Nucleic Acid Probes
|
For ordering of Locked Nucleic Acid Probes please follow these instructions. For any help please contact the biomers.net customer support.
|
 |

