
Synthesis of Oligonucleotides
Automated synthesis of oligonucleotides
Advanced synthesis chemistry and state-of-the-art parallel DNA synthesisers allow today for a fast and efficient production of oligonucleotides of almost all sequences. In a so-called solid-phase synthesis the desired oligonucleotide sequence is synthesised onto an inert solid support, from which it is cleaved after the synthesis has been completed. In comparison to a synthesis in liquid phase, the solid-phase synthesis has the big advantage of not having to isolate the different intermediate products - a tedious and time consuming work - but being able to remove unused reagents and certain by-products by applying simple washing and filtration steps.
Synthesis
Synthesis
Raw materials
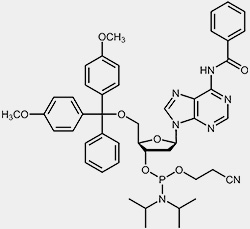
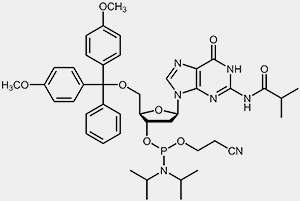
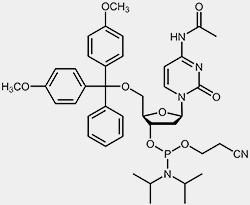
The most important raw materials for the oligonucleotide synthesis are the so-called nucleoside phosphoramidites.
| 2'-deoxy-adenosine-phosphoramidite | 2'-deoxy-guanosine-phosphoramidite |
|---|---|
 |
 |
| 2'-deoxy-cytidine-phosphoramidite | 2'-deoxy-thymidine-phosphoramidite |
|---|---|
 |
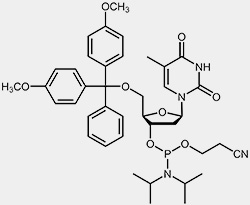 |
The reactive phosphoramidite group is located at the 3’-OH, the 5’-OH is blocked with the acid-labile dimethoxytrityl protection group (DMT). To prevent the exocyclic amino-functions of adenosine, guanosine and cytidine from undergoing unwanted side-reactions they have been blocked using acyl protecting groups.
To start the synthesis, a solid support that has been derivatised with the respective 3'-nucleoside of the oligo to be synthesised is required. Usually the starting nucleotide is bound to controlled pore glass (CPG) or polystyrol beads by a base-labile linker:
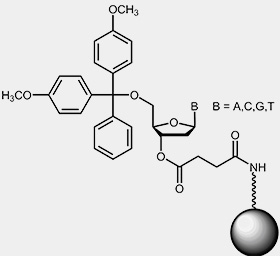 |
Synthesis cycle
The chemical synthesis of oligonucleotides is performed from 3' to 5'. During the key coupling step a 3'-phosphoramidite reacts with a free 5'-hydroxy group:
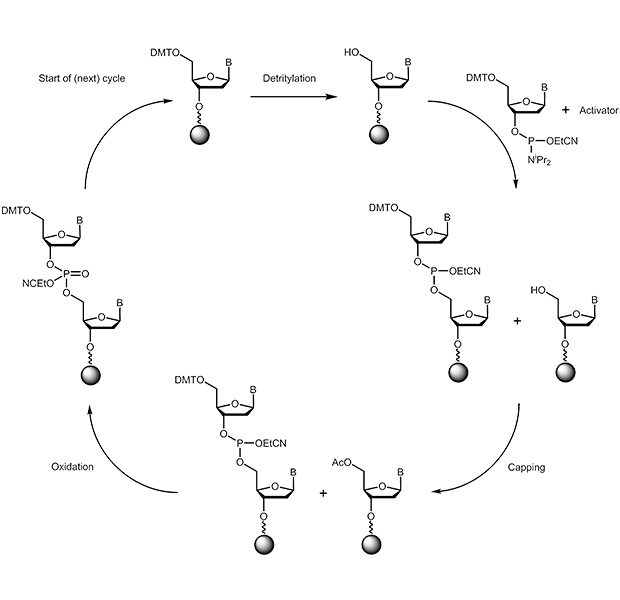 |
a) Deblocking:
The acid-labile 5’-dimethoxytrityl protecting group is cleaved from the base that is anchored to the solid phase support (either start nucleoside or later the growing oligo strand); thereby a free reactive hydroxy function is obtained. As cleavage reagent di- or tri-chlor acetic acid in dichlormethane is used.
b) Coupling:
The free 5’-OH group is now able to react with added phosphoramidite. As a result both nucleosides are linked by a phosphit bridge. The phosphoramidite has to be activated first using a weak acid (e.g. 1H-tetrazole).
c) Capping:
As the coupling step, as every chemical reaction, is not to 100% effective, one has to prevent remaining free OH-groups (about 1%) from reacting in the following synthesis cycle, thus creating non-specific sequences. Therefore in the capping step all free reactive groups are blocked using acetylation, thus eliminating them as reactive partners in the further course of the synthesis.
d) Oxidation:
The internucleotide phosphit group that has been created in the coupling step is oxidised to its phosphate using iodine solution.
A new synthesis cycle is started from step a) again. These reactions are repeated until the desired oligonucleotide sequence has been produced. The cycle can be terminated to result in either a solid-support-bound oligonucleotide carrying a free 5'-OH group or one carrying a protected 5'-OH group (DMT).
After accomplishing the synthesis post-processing steps are required
a) Cleavage of the oligo from the solid support:
By treating the support-bound oligonucleotide with concentrated ammonia solution, the ester bond between the 3'-OH of the oligo and the solid support is cleaved, the oligo now "swims" in ammonia solution.
b) Cleavage of the base protecting groups:
To remove the protecting groups from the adenine, guanine and cytidine bases and to release the exocyclic amino function, the ammonia oligonucleotide solution is incubated at higher temperatures (usually between 50°C and 80°C for 1 to 8 hrs. depending on the protocol and protecting groups used).
Stepwise coupling yields
Stepwise coupling
Stepwise coupling yields in DNA synthesis refers to the efficiency of coupling a new phosphoramidite to the growing oligonucleotide strand, thus the yield over the steps of a reaction cycle.To form a reactive 5' OH, at the beginning of each coupling cycle the dimethoxytrityl protection group is cleaved by acid treatment. The amount of released dimethoxytrityl cation can be determined by absorption or conductivity measurement. Assuming a total cleavage of DMT groups from the oligonucleotide strand one can comprise from the amount of DMT cation the efficiency of the previous coupling step. These 'trityl values' and therefore the stepwise coupling yields are a measure for the quality of the synthesis process. The average stepwise coupling yield during an oligonucleotide synthesis has a drastic impact on the percentual amount of target sequence that can theoretically be obtained:
% Target sequence = (average stepwise coupling yield) n (n=number of bases)
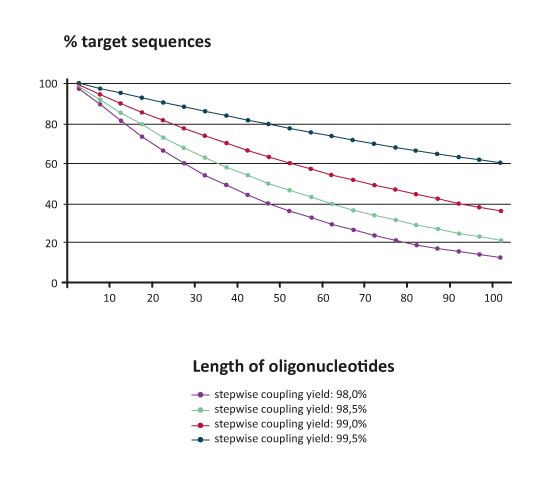 |
The diagram above shows that e.g. an 80mer raw product with an excellent average coupling yield of 99% will contain calculationally clearly below 50% of target sequence; the rest consists of shorter n-x products.Therefore, depending on the planned experiment one has to consider carefully which purification method should be used for further workup of raw oligonucleotides.
Deprotection
Deprotection
During the deprotection process the oligonucleotides are cleaved off from the solid support where the synthesis took place. During the same step the base protecting groups and the protecting groups on the phosphate backbone are removed as well. These protecting groups prohibited side reactions during the synthesis cycles. The terminal trityl protecting group is usually removed after the purification of the oligonucleotides.
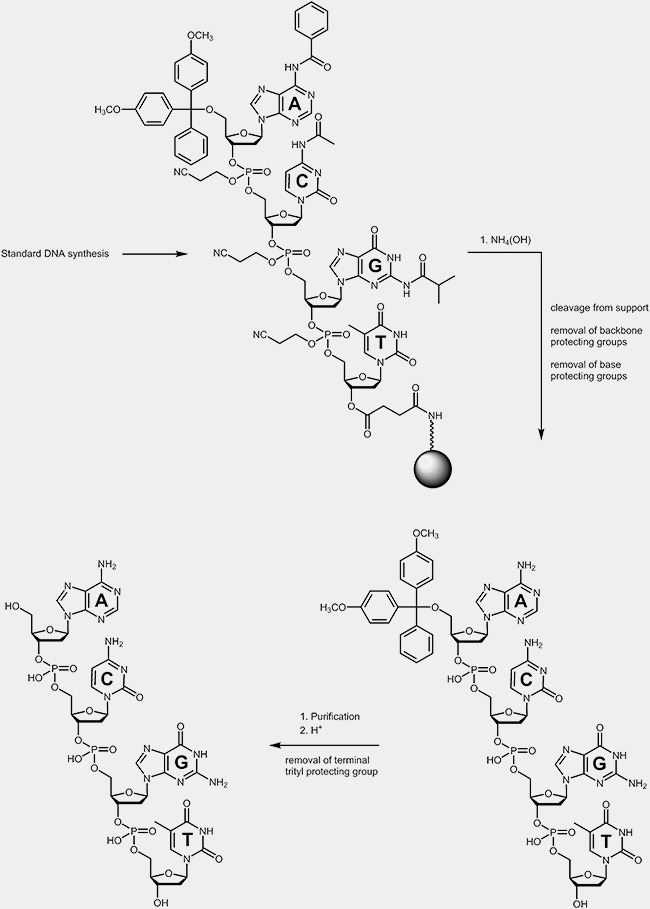 |

