
Purification
Below find details to the different possiblities biomers.net offers to purify an oligo. In some cases, a particular purification is the default stage (e.g.: HPLC for dye labelled oligos); in other cases it may depend on the experimental set-up and needs, which purification is sufficient or neccessary. To form your own opinion, please find here informations for the different purifications.
Cartridge
RP (reverse phase) cartridge
Standard purification
Following the synthesis of the oligonucleotide the hydrophobic dimethoxytrityl protection group is left at the 5´-end of the oligonucleotide (DMT on). This is the criteria to differentiate between the full length product and the by-products, which mainly consists of shorter sequences. After deprotection the liquid solution of the raw synthesis product is given onto the RP-cartridge. Those oligonucleotide comprising the full length product (DMT on) are strongly absorbed by the cartrigde material, while the shorter fragments missing the final DMT-protection group are easily washed away.
By several washing steps the shorter fragments and potential salts are eliminated. Finally, the last protection group is separated from the purified oligonucleotide by an acidic treatment, enabling the elution of the desired product with a water/acetonitril mixture. The result is a purified, saltfree oligonucleotide.
According to experience, the purification obtained with this methods is in the range of short unmodified oligonucleotides (up to 30 bases) quite comparable to that purification observed after HPLC. All our oligonucleotides (up to ~40 bases) are controlled by Maldi spectrometry subsequently to the purification.
RP HPLC
HPLC purification for oligos
For longer oligonucleotides (> 30 bases) a purification with reverse phase HPLC is recommended. Due to the distinct choice of material and length of the cartridge and the elution gradient an optimal separation of product and unwanted artefacts can be obtained. Here also the discrimination is achieved by the final DMT protection group left on the full length oligonucleotide. The purification process is controlled and documented by UV spectroscopy. This enables the concentration of the pure product by collection of the desired peak , which is „cut for quality“, so that any peak shoulders will not be collected.
For modified oligonucleotides the RP HPLC also is the method of choice. Particularly for dye modified oligonucleotides the purification is controlled by measurement of two wavelength, one at 260 nm to detect the bases and an additional that detects in the absorption maximum of the dye.
Dual labelled probes and Molecular Beacons are subjected to a specially adapted purification process with several steps.
IEX-HPLC
IEX-HPLC
The principle of ion exchange chromatography is based on interactions between contrary charged ions. This method enables the separation of species with a different number of charges from each other. For the purification of oligonuncleotides with their negatively charged phosphate backbone, anionic ion exchange is the method of choice. As stationary phase a resin with positively charged functional groups is used. These then interact with the negative charges of the oligonucleotides. The separation of strands with N nucleotides of failure sequences with N-1, N-2, and so forth nucleotides is induced by a gradient of increasing ionic strength. Strands with lesser negative charges will elute first while the longer product strands are still retained and will elute in a final step. During the last years the importance of ion exchange chromatography as an alternative and additional purification method has increased steadily.
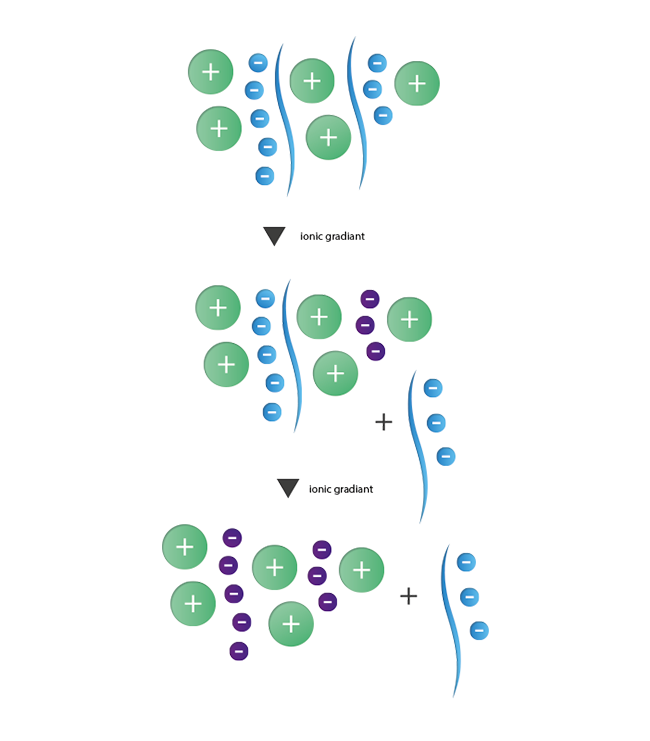 |
PAGE
Polyacrylamide gel electrophoresis (PAGE)
In the purification methods described so far, the criterion to differentiate between desired and unwanted products is the DMT protection group. With PAGE purification the separation is done according to charge and the molecular weight of the oligonucleotide.
The completely deprotected oligonucleotides are segregated under denaturing conditions on acryl amid gels of different composition. This allows in optimal cases the differentiation of oligonucleotides which vary in just one base length.
After electrophoresis, the bands are visualised by „UV shadowing“ and the desired band will be cut from the gel. With water or buffer the oligonucleotide is eluted from the gel matrix and subsequently is desalted and quantified.
In particular, long oligonucleotides subjected to cloning or mutagenesis experiments are prominent candidates
for PAGE purification.

