
Enzymatic coupling via oligonucleotides with
protein tag-specific ligands
Introduction
Enzymatic coupling via oligonucleotides
Studying the structure and function of proteins has been a high priority in research for years. Using recombinant DNA systems, proteins can be easily expressed and isolated in foreign species.
Some protein tags have proven to be very promising. So-called epitope affinity tags bound to the desired protein enable selective purification of recombinant proteins from the cell extract. These include, for example, HisTag® (poly-histidine), GST tag (glutathione S-transferase), MBP tag (maltose binding protein), CBP tag (calmodulin binding protein) or FLAG tag. Using affinity chromatography can achieve an increase in protein purity and yield. 1
Protein tags of the next generation, such as SNAP or CLIP tag as well as HaloTag®, are characterised by a much wider range of applications and are used in various fields:
- detection
- localisation
- protein-protein interactions
- labelling, multiplex labelling (e.g. SNAP / CLIP tag)
- enzymatic coupling of different proteins or other biomolecules
With biomers.net you can order oligonucleotide ligands for a variety of protein tags (HaloTag®, SNAP tag, CLIP tag, etc.).
For further analysis of the fusion proteins, the oligonucleotides are labelled with tag-specific ligands.
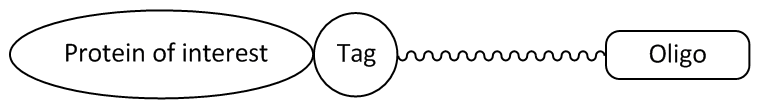
Picture 1: Oligonucleotide ligand with a tag-specific linker that covalently binds the fusion protein.
Protein tags of the next generation are generally small proteins and can be attached to a target protein by translation.
The function of these protein tags, such as the HaloTag® or the SNAP or CLIP tag, is based on the recognition of a particular substrate or ligand (e.g. fluorescence conjugate of an oligonucleotide and a fluorescent dye). The ligand can be recognised highly specifically by the tag and covalently bound, whereupon further studies of the fusion protein are possible.
By exchanging the oligonucleotide ligand, the function as well as the properties of the fusion protein can be influenced (e.g. membrane permeability, fluorescence labelling, surface binding, etc.).
Literature:
1. Overview of Affinty Tags for Protein Purification. Kimple ME, Brill AL, Pasker RL; Curr Protoc Protein Sci. (2015); 73,: Unit-9-9.
2. Snap-, CLIP- and Halo-Tag Labelling of Budding Yeast Cells. Stagge F, Mitronova GY, Belov VN, Wurm CA, Jakobs S; Plos One (2013); Volume 8, Issue 10, e78745.
3. Motor reattachment kinetics play a dominant role in multimotor-driven cargo transport. Feng Q, Mickolajczyk KJ, Chen G-Y, Hancock WO; CC-BY-NC-ND 4.0 International license.
SNAP-/CLIP-Tag
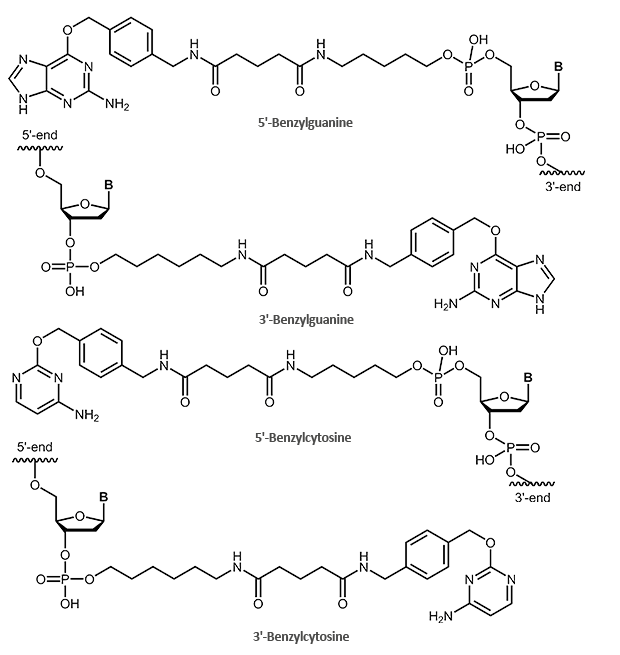
SNAP and CLIP tag protein ligands – benzylguanine- and benzylcytosine-labelled oligonucleotides
There are a variety of possible tools that allow selective protein labelling in living cells. In addition to the HaloTag®, the SNAP tag and the CLIP tag have been particularly highlighted. Both tags allow the visualisation of complex, cellular processes (e.g. protein-protein interactions, protein localisation, protein function analyses). Here, too, the protein tag is fused to the desired "protein of interest" by means of a vector expression system. The tag can be bound to the N- or C-terminal of the target protein.
The SNAP tag itself is a small protein derived from the 20 kDa human DNA repair protein O6-alkylguanine DNA alkyltransferase (AGT). The substrate is the O6-benzylguanine (BG) derivative.
In this process, the benzyl group of the BG derivative is transferred to the cysteine in the active site of the protein tag, so that tag and BG derivative are covalently linked. The guanine group is released.
A modified form of the SNAP tag is the CLIP tag. This tag is also based on the enzyme AGT, but reacts specifically with O2-benzylcytosine (BC) derivatives.
Due to the orthogonal substrate specificity and the lack of cross-reactivity between the two tags, a so-called multicolour labelling is possible. Here, SNAP and CLIP tag fusion proteins can simultaneously be used for labelling of multiple proteins in a cell.
Oligonucleotides with SNAP and CLIP tag ligands
With biomers.net you can label your oligos at the 5'- or 3'-end with a suitable protein tag ligand. For this purpose, the oligo is either labelled with a benzylguanine (SNAP tag) or a benzylcytosine (CLIP tag). The oligo itself can also be further modified (fluorescent dyes, biotin).
Literature:
1. An Engineered Protein Tag for Multiprotein Labeling in Living Cells. Gautier A, Juillerat A, Heinis C, Reis Corrêa I, Kindermann M, Beaufils F, Johnsson K; Chemistry and Biology 15 (2008), 128-136, DOI 10.1016.
2. Simultaneous Protein Tagging in Two Colors. Schultz C, Köhn M; Chemistry & Biology (2008), 91-2. doi: 10.1016/j.chembiol.2008.02.004.
Sortase/Triglycine
Sortase-mediated coupling reaction via triglycine-modified oligos
In general, sortases are bacterial, membrane-associated enzymes and belong to the class of transpeptidases. In addition to the structural formation of bacterial pili, their function encompasses the anchoring of secreted proteins from the cytoplasm into the cell wall envelope of Gram-positive bacteria. 1.2
In addition to structuring proteins, important virulence factors are also integrated into the cell wall, which play a critical role in bacterial pathogenesis. The inactivation of involved sortases can influence or even inhibit the virulence of bacteria.3,4
For several years, the transpeptidase reaction of the sortase has also been applied in vitro:
- covalent coupling of different proteins or other biomolecules
- analysis of the binding properties of a protein, e.g. by means of dye-labelled antibodies or ligands
- PEGylation of molecules in therapeutics: coupling of polyethylene glycol (PEG) to a biomolecule prevents its intracellular degradation and thus increases the pharmacodynamic properties 5
- cyclisation of proteins and thus inhibition of degradation 6
The compound is based on a specific terminal recognition motif on the target protein – the LPXTG motif (leucine-proline-Xaa-threonine-glycine). X stands for any amino acid. Sortases are able to cleave the LPXTG sequence on the peptide to link it subsequently to the desired ligand (oligonucleotide, peptide, protein, etc.).
In the first step, an intermediate product of sortase and target protein is formed. For this purpose, the thiol residue of the cysteine in the active site of the sortase binds the threonine within the recognition sequence. The C-terminal glycine residue is cleaved off. In the next step, the carboxylic group of the threonine binds the amino group of the free terminal triglycine, so that target protein and biomolecule are covalently linked.7
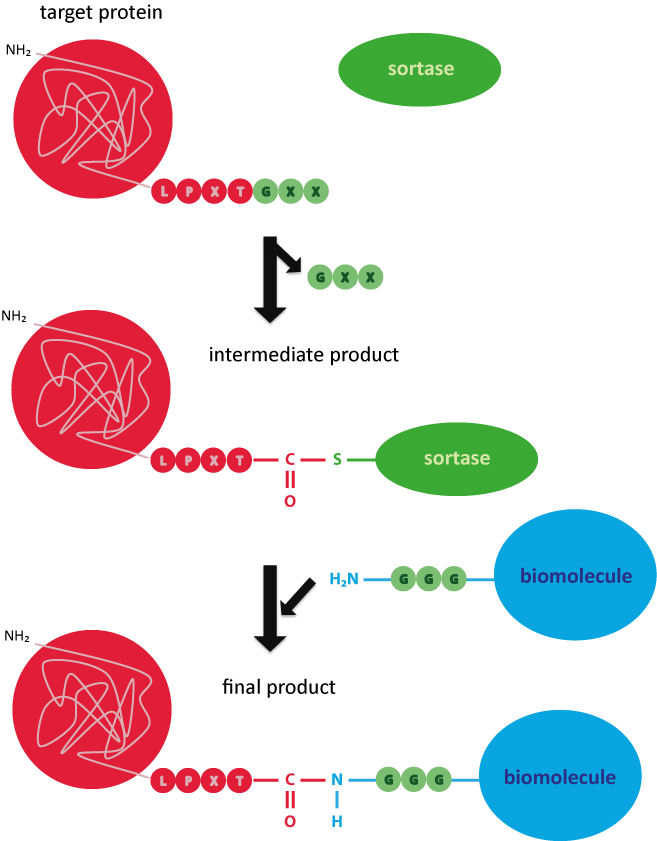
Picture 3: Reaction mechanism of the sortase for covalently coupling a target protein to a triglycine-labelled biomolecule.
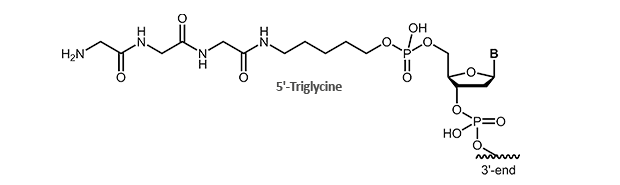
For this application we offer triglycine-modified oligonucleotides. The modification is available for the 5´-end of the oligonucleotide.
For further information you can contact us at any time!
Literature:
1. Sortase Transpeptidases: Insights into Mechanism, Substrate Specificity and Inhibition. Clancy KW, Melvin JA, McCafferty DG; Biopolymers (2010), 94(4): 385–396. doi: 10.1002/bip.21472.
2. Sortase enzymes in Gram-positive bacteria. Spirig T, Weiner EM, Clubb RT; Mol Microbiol. (2011), 82(5):1044-59. doi: 10.1111.
3. Sortase A: an ideal target for anti-virulence drug development. Cascioferro S, Totsika M, Schillaci D; Microb Pathog. (2014), 77:105-12. doi: 10.1016.
4. Inhibition of sortase-mediated Staphylococcus aureus adhesion to fibronectin via fibronectin-binding protein by sortase inhibitors. Oh KB, Oh MN, Kim JG, Shin DS, Shin J; Appl Microbiol Biotechnol. (2006), 70(1):102-6. Epub 2005 Jul 12.
5. Sortase-catalyzed transformations that improve the properties of cytokines. Popp MW, Dougan SK, Chuang T-Y, Spooner E, Ploegha HL; Proc Natl Acad Sci U S A (2011), 108(8): 3169–3174.
6. Sortase-mediated backbone cyclization of proteins and peptides. van 't Hof W, Hansenová Maňásková S, Veerman EC, Bolscher JG; Biol Chem. (2015), 396(4):283-93. doi: 10.1515/hsz-2014-0260.
7. Sortase-mediated protein ligation: a new method for protein engineering. Mao H, Hart SA, Schink A, Pollok BA; J Am Chem Soc. (2004), 126(9):2670-1.

