
Reactive Linkers
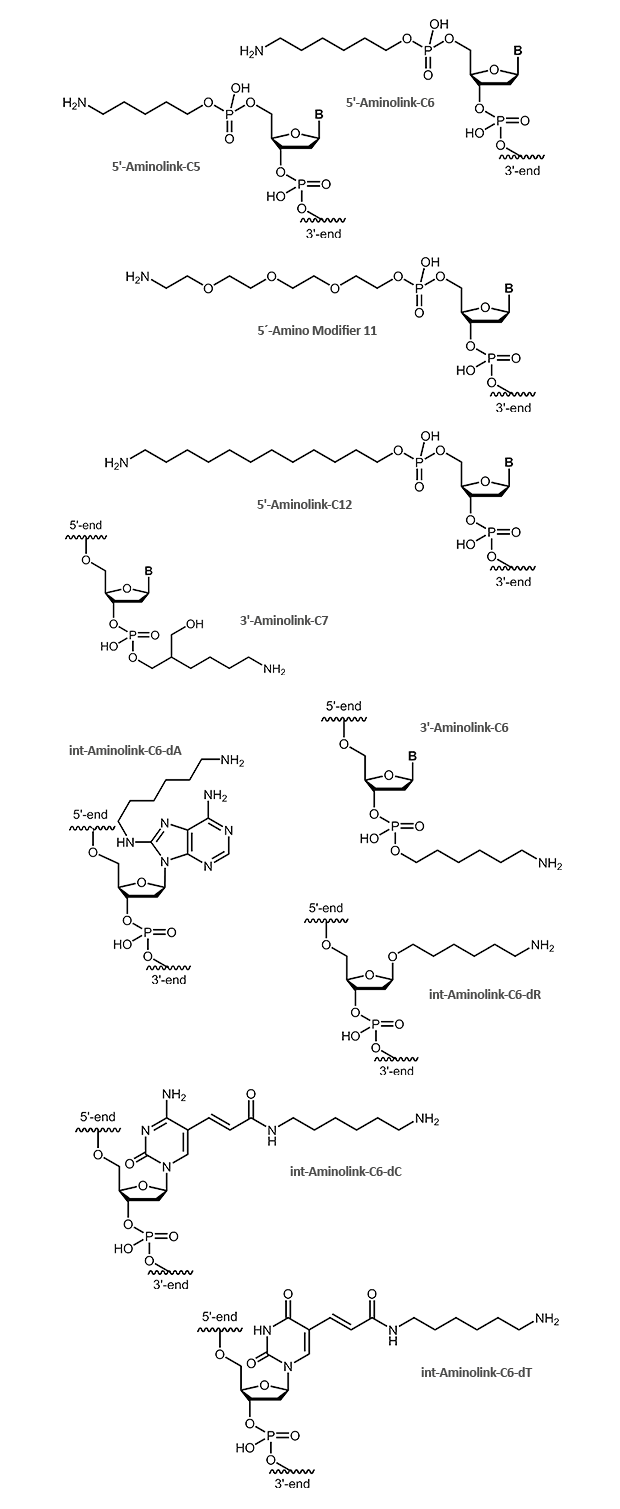
Aminolink
Aminolink
An aminolink adds a terminal amino group (NH2) bound to a linker. The amino group can be used to couple additional molecules, e.g. dyes or proteins like HRP. The aminolink is used as well with DNA arrays to bind oligonucleotides to adequately prepared surfaces. There are linkers of variable length for use in different applications. Aminolinks can be coupled to both the 5´- and the 3´-end of the oligonucleotide. Modifications at the
3´-terminus of oligonucleotides make them more resistant against exonuclease digestion. Furthermore, it can be useful to link an aminogroup inside the oligonucleotide. For this purpose, a thymidine nucleotide's C5 methyl group can be easily replaced by a C6 linker with an amino group at its end. Thus the interaction between the amino group and the DNA is reduced as far as possible so that the modified oligonucleotide behaves in hybridisation comparable to a respective unmodified one.
The figures below show the deprotected final product as it will be finally delivered with the oligonucleotide.
 |
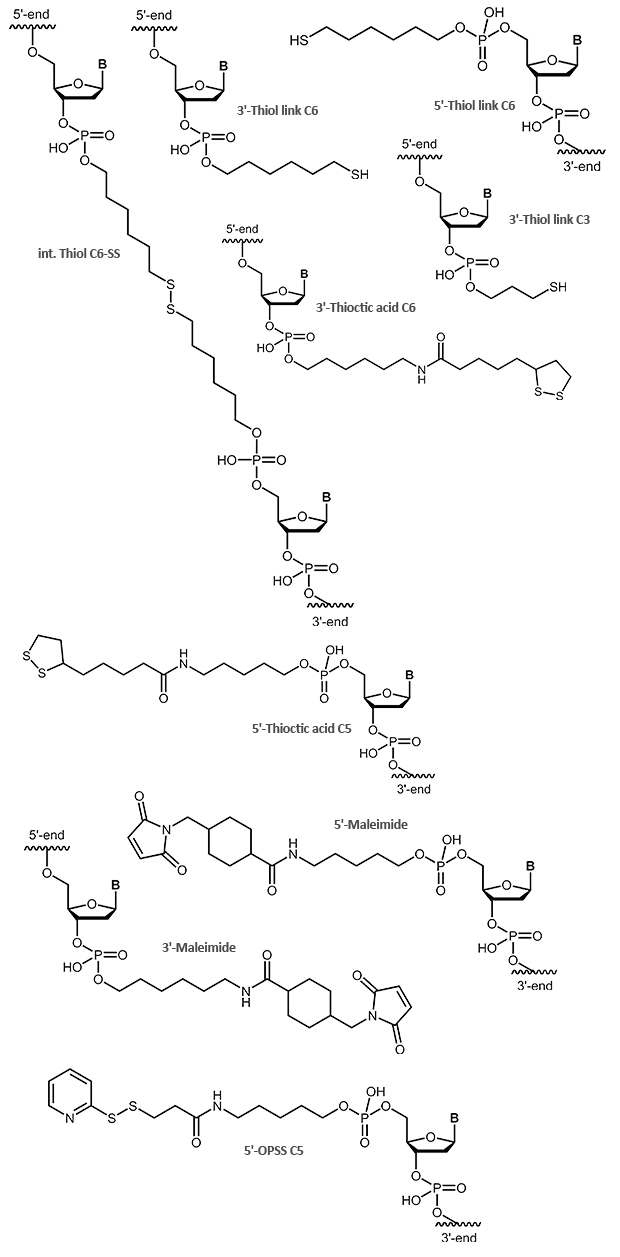
Thiol / Maleimide
Thiol and Maleimide
Sulfur compounds are often used to bind molecules to gold particles. Like aminolinks, thiol links at the 5´-end of an oligonucleotide are linked to the respective end of the oligonucleotide using a chain of 6 carbon atoms (6xCH2). At the 3´-end a SH group is linked using a C3 linker. Thiol links are frequently used to bind an oligonucleotide as a thioether covalent to maleimides. Thiol and maleimides are easily reacted forming a stable covalent linkage with each other. Dependent on the reactive agent available for the modifier, the oligo can either be activated with a thiol or a maleimide function, both options are available. This offers various means to couple an oligonucleotide that has been modified in such a way to e.g. dyes or proteins.
In addition to a simple thiol, a thioctic acid modification is also suitable for binding oligonucleotides to gold surfaces. Due to the two sulfur atoms the bond to the gold is even stronger.
OPSS, the orthopyridyl disulfide, is a terminal modification that can form a stable disulfide bridge with a SH group (e.g. cysteine). Here, proteins, peptides or other biomolecules, as well as surfaces, can be coupled to an OPSS-modified oligonucleotide.
 |
Click Chemistry
Click Chemistry
The term „click chemistry“ describes a fast and thermodynamically favoured reaction which enables an efficient and selective linkage of two molecules. In a more specific sense the click reaction is a cycloaddition between an azide and an alkyne either under copper catalyzed or copper free reaction conditions. Due to the fact that such click reactions work efficiently in aqueous media, they are very much suitable for modifying biomolecules or linking different biomolecules together. Furthermore the azide and alkyne reaction partners do not interfere with other functional groups like e.g. amino or carboxy which opens an additional degree of freedom for orthogonal coupling strategies.
A selection of alkine and azide linker as well as the copper-free variants (DBCO, TCO, Tetrazine) can be found at Click Chemistry.
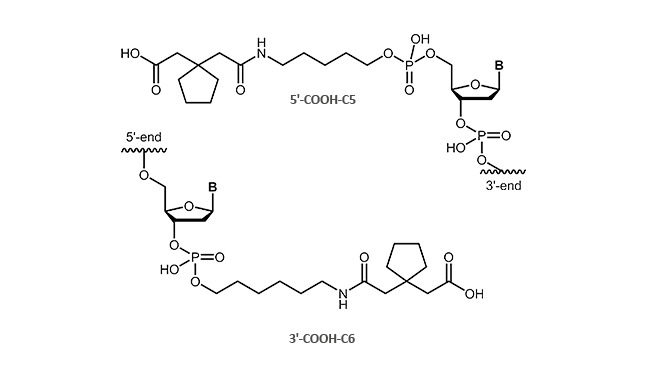
Carboxylinker
Carboxylinker
 |
Aldehyde
Aldehyde
Aldehydes are highly reactive molecules that can be used to link oligonucleotides to other biomolecules. In general, carbonyl compounds (aldehydes) react with nucleophiles (amino groups, thiol compounds, etc.). The amino groups can be simple amine linkers (primary and secondary amines), but also hydrazine, hydrazide or aminooxy compounds can react with aldehydes.
Due to the partly complex reactions with aldehydes, reactions with aminooxy compounds (e.g. aminooxyacetic acid) are preferred. The aminooxyacetic acid (AOA) reacts with aldehydes to form stable oximes.
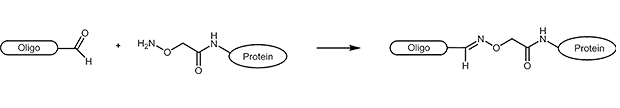
Picture 1: Reaction scheme of an aldehyde-labelled oligonucleotide and an AOA-coupled protein.
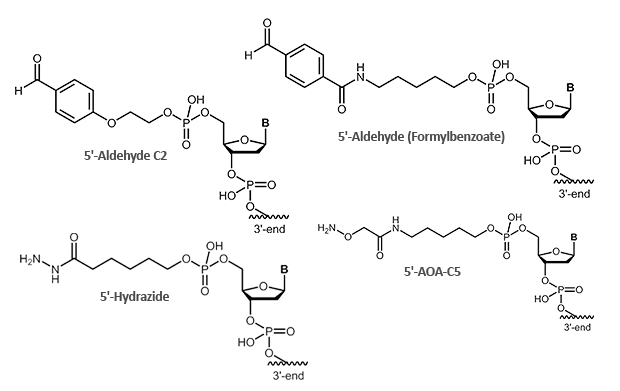 |
Literatur:
1. Organic Chemistry, 4th edition. Carey FA; Chapter 22, p. 858.
CBT / Cysteine
Cyanobenzothiazole and Cysteine – effective protein labelling with modified oligonucleotides
An efficient approach to combine different biomolecules is presented by the conjugation of cyanobenzothiazole (CBT) and aminothiols as for example cysteine. The conjugate of these two molecules is long known as the last step in the generation of luciferin, the light-generating substrate of the enzyme luciferase. 1 The reaction of aminothiols and cyanobenzothiazole proceeds spontaneously at physiological conditions, even inside living cells. 2
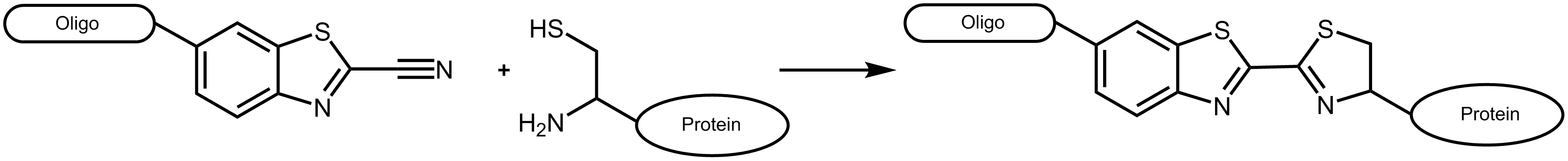
Figure 1: Reaction scheme of a cyanobenzothiazole-labelled oligonucleotide with a suitable protein carrying an N-terminal cysteine.
Cyanobenzothiazole reacts with cysteines exclusively at the N-terminal position of a protein, not with cysteines inside the amino acid sequence (for an exception see Ramil et al. 3). As naturally occurring proteins usually have no terminal cysteine, this allows an efficient and specific labelling by respectively engineered proteins.
We offer cyanobenzothiazole as a 5´-modification for oligonucleotides up to a length of 40 bases.
Cyanobenzothiazol can react as NHS ester with an oligonucleotide bearing a primary amino function [chemical structure] or as azide via click chemistry with an alkyne linker [chemical structure].
Besides coupling to proteins (see figure 3a), the CBT-cysteine-reaction is also used for the assembly of nanostructures 2 or for immobilisation 4. Thus, besides CBT-modified oligonucleotides, also cysteine can be attached to the 5´-end of oligos (figure 3b, exemplarily conjugation of two oligonucleotides).
a) b)
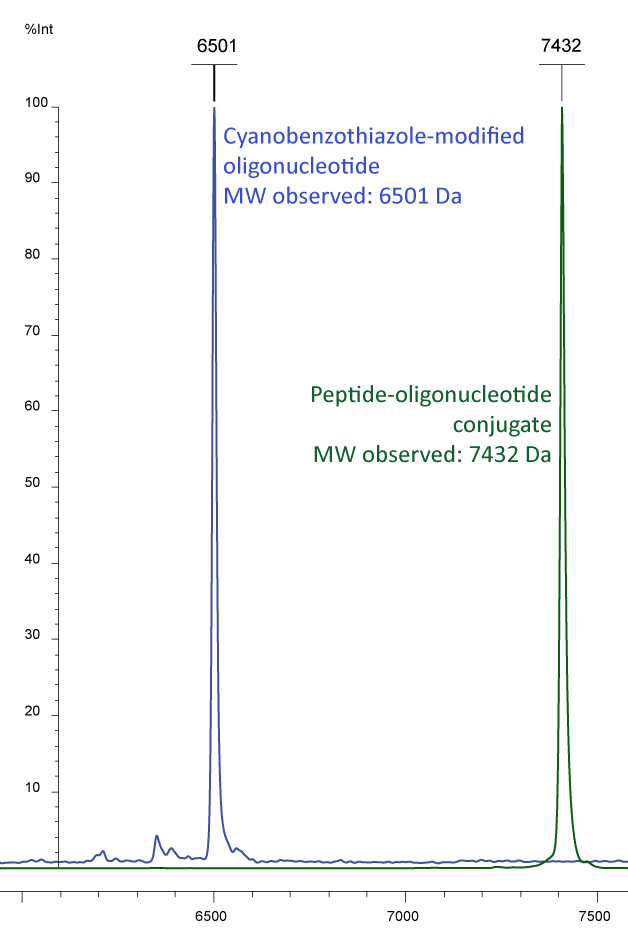
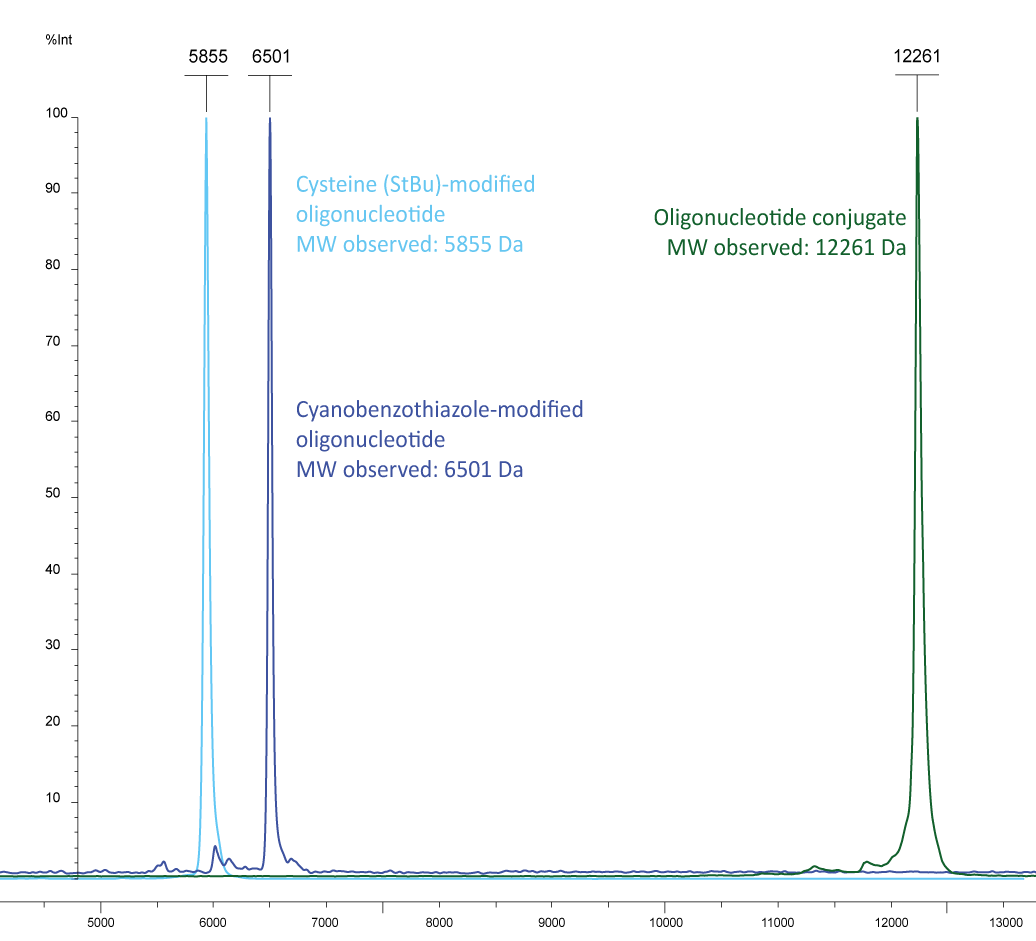
Figure 3a) shows the Maldi analyses of a conjugation study of an oligo and a protein. The peak in blue shows a 5´-cyanobenzothiazole oligo with an additional fluorescent modification at the 3'-end: EUB 338-probe CBT-gctgcctcccgtaggagt-6-Fam (MW 6497). This oligo was conjugated with a poly-histidine, bearing an N-terminal cysteine (MW 935). The result of the conjugation is presented by the green peak, showing the sum of the molecular weights.
Figure 3b): In light blue a cysteine labelled oligonucleotide (5´-cysteine-gctgcctcccgtaggagt, MW 5849) and in dark blue a 5´-cyanobenzothiazole-EUB 338-probe (5´-CBT-gctgcctcccgtaggagt-6-Fam, MW 6497) are shown, the conjugate is shown in green with the molecular weight of 12261, representing the sum of the two individual molecules.
Please note that the cysteine (StBu protecting group) is supplied protected and needs to be treated with an appropriate reducing agent (e.g. TCEP) before further conjugation steps, in order to reduce the disulfide and release the thiol group. The TCEP can be added, for example, to the conjugation buffer.
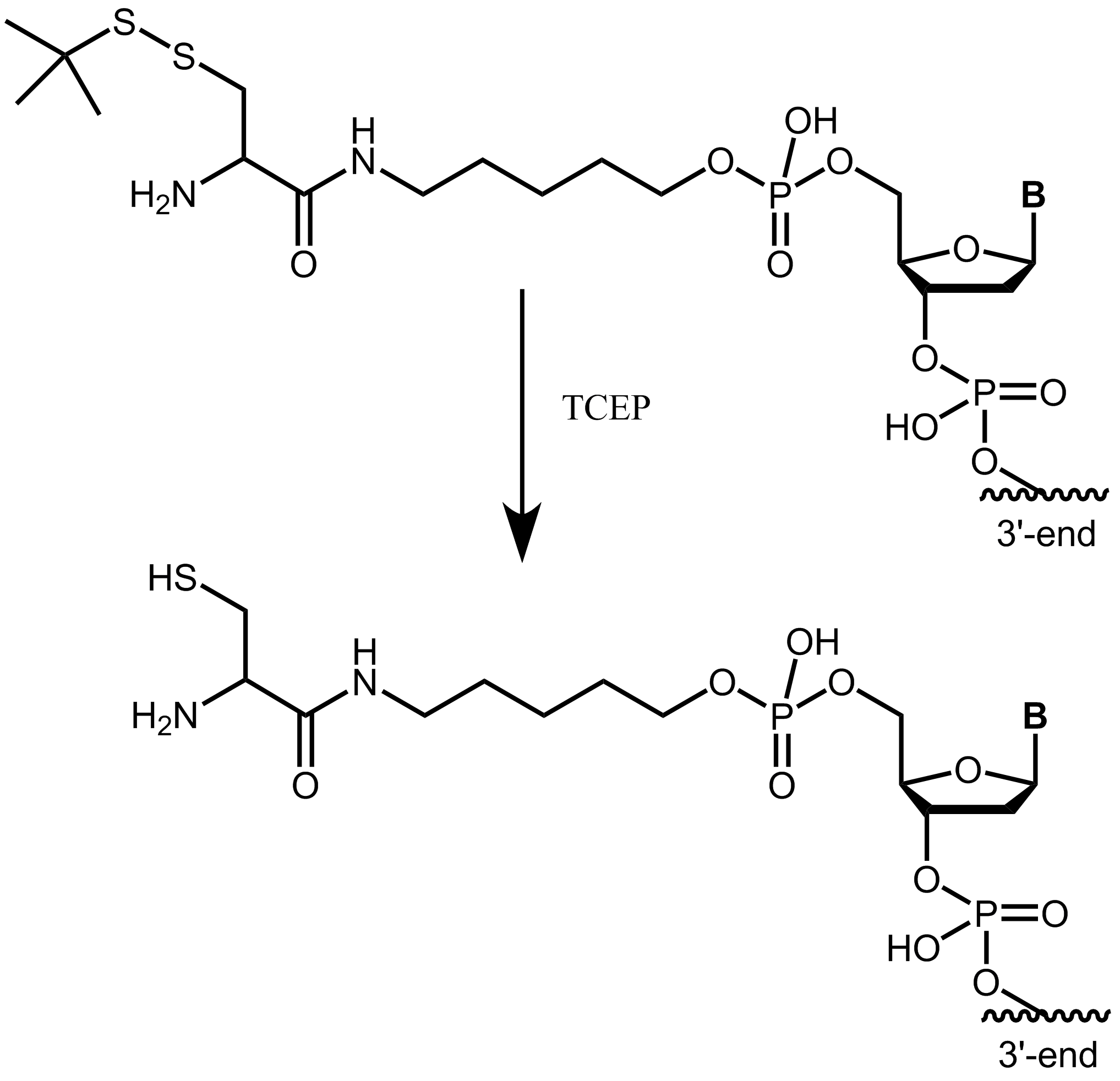
Figure 4: Cysteine with StBu protecting group requires treatment with TCEP to obtain free, reactive cysteine.
Literature:
1. The structure and synthesis of firefly luciferin. White EH, McCapra F, Field GF, McElroy WD; J. Am. Chem. Soc. (1961), 83:2402–2403.
2. A biocompatible condensation reaction for controlled assembly of nanostructures in live cells. Liang G, Ren H, Rao J; Nat Chem. (2010), 2(1): 54–60.
3. Sequence-Specific 2-Cyanobenzothiazole Ligation. Ramil CP, An P, Yu Z, Lin Q; J Am Chem Soc. (2016), 15(6):829-35.
4. Site-specific immobilization of biomolecules by a biocompatible reaction between terminal cysteine and 2-cyanobenzothiazole. Wang P, Zhang CJ, Chen G, Na Z, Yao SQ, Sun H; Chem Commun (Camb). (2013); 49(77):8644-6.
Photoreactive linkers
Photoreactive linkers - Photocleavable linkers and Photocrosslinkers
The specific release of a molecule as well as the coupling of two biomolecules at particular time is an extremely desirable feature in many applications (e.g. bioconjugation). An external, non-invasive pulse (here light) can be used for controlled cleavage (Photocleavable linker) or coupling (Photocrosslinker) so that the biomolecules can be released or linked at particular time.
Generally, photolabile linkers are considered as a connection between two molecules (oligonucleotide sequences, peptides, etc.) via a short linker which can be activated and subsequently cleaved after irradiation with light of a specific wavelength (UV light at 300-400 nm).
Photoreactive linkers are highly selective and show high stability in different chemical conditions. Here, the light provides a simple, well-regulated "switch function" in conjugations, as it allows a specific, temporally controllable reaction (cleavage or coupling of biomolecules).
Photocleavable linkers – light-induced cleavage of labelled oligonucleotides
biomers.net offers different UV-light-sensitive linker structures:
- PC Linker
- PC Spacer
- PC Biotin
- PC Aminolink C6
In addition to the internal modifications PC Linker and PC Spacer, also cleavable, terminal modifications, such as
PC Biotin and the PC Aminolink C6, are possible. The internal modifications PC Linker and PC Spacer can be inserted at any position within your oligonucleotide sequence.
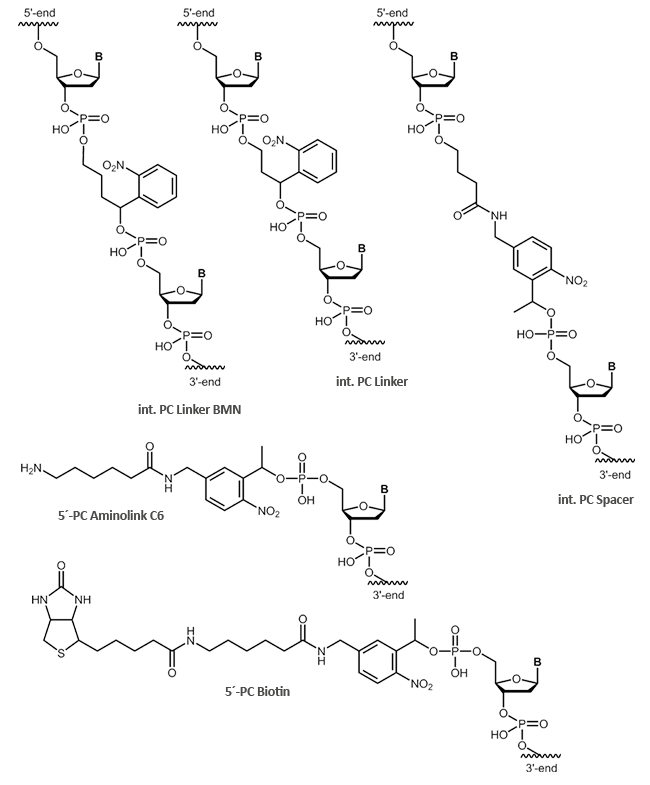
Using photocleavable linkers (PC linkers), organic substrates can be coupled to solid phases or other biomolecules. Especially in so-called photo-induced hybridisation assays, PC linkers can be used efficiently.
For localisation and structural characterisation of biomarkers, so-called Tag-Mass probes are used. The Tag-Mass is based on the indirect detection of mRNA. In this case, the specific probe binds to a target sequence and the presence of the mRNA can be detected indirectly via a coupled, cleavable tag of known mass (specific Maldi imaging). The Tag-Mass is bound to the target-specific probe via a PC linker. Since the wavelength for separation is close to that of the actual Maldi analysis, the tag is released and can be detected during the Maldi measurement. Tag-Mass thus allows a rapid and highly sensitive measurement of mRNA and protein expression levels, thereby providing new information for transcriptome/proteome colocalisation studies.
In addition, Maldi imaging allows rapid detection and localisation of biomolecules in tissue, so that time-consuming extraction, purification and separation are no longer necessary.
The most commonly used PC linkers are based on a nitrobenzyl group. A light pulse in the range of 300-400 nm can cleave the N=O double bond of the nitro group, resulting in a diradical excited state. In further steps of this reaction, the diradical state can be finally converted into a stable 2-nitrosobenzaldehyde. The duration of light exposure can regulate the amount of cleaved biomolecules.
Photocleavable linkers allow a high degree of chemoselectivity by controlling spatial, temporal, and concentration-dependent variables through the addition of light. Since the removal of the linker structure requires no chemical additives, this method is often referred to as a "traceless" process.
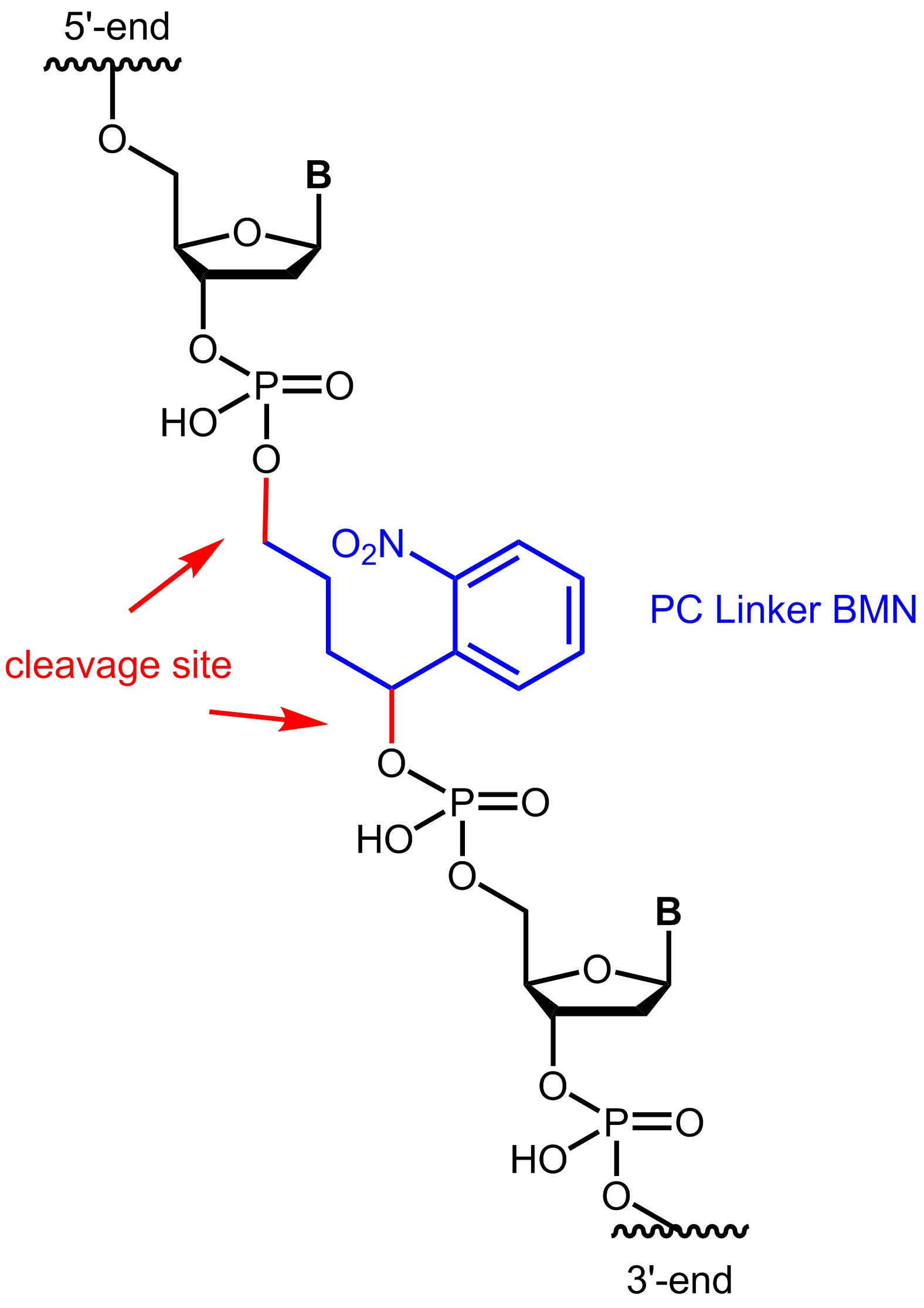
Figure 1: Structure of the internal photocleavable linker BMN (PC linker BMN) bound to an oligo.
Even the short exposure to high-energy light using a Maldi measurement is sufficient to display the decomposition products of an oligo with an internal PC linker.
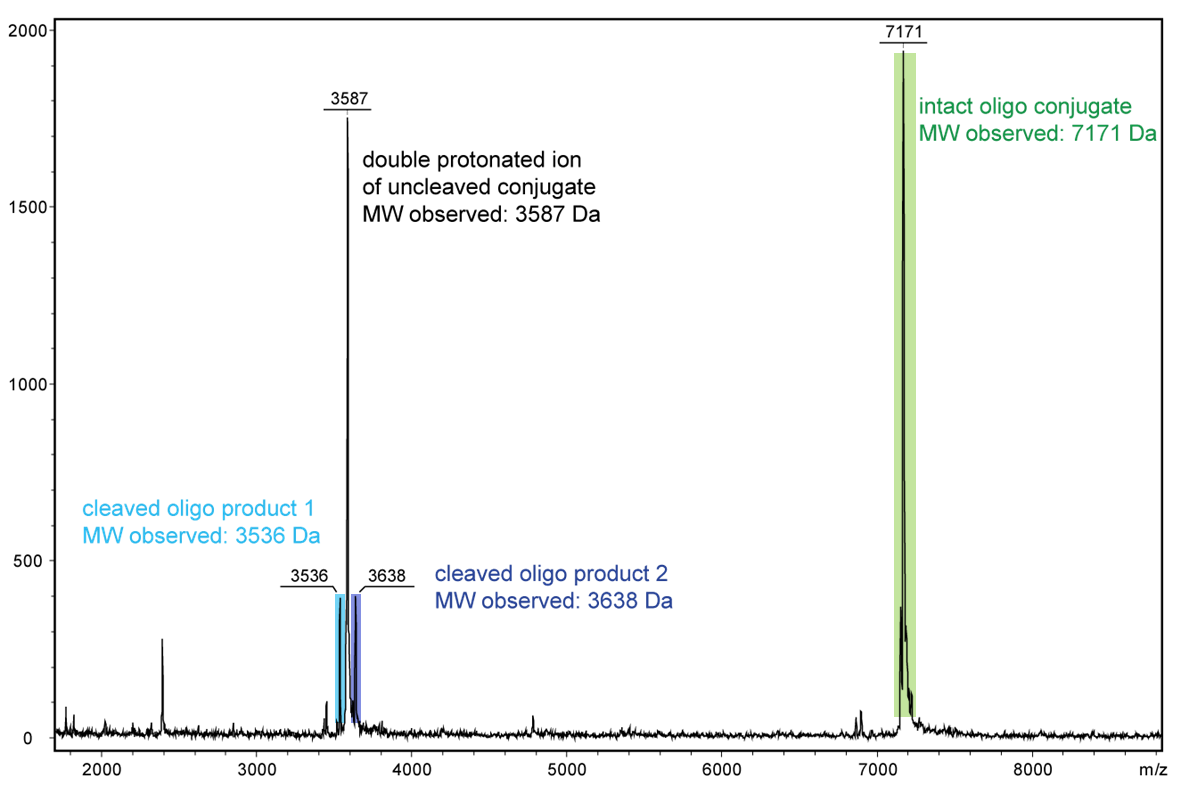
Figure 2 shows the result of the Maldi analysis of a dT20 oligonucleotide with an internal photolabile linker (PC). The complete oligonucleotide with a molecular weight of 7171 Da is shown in the large peak on the right side (highlighted in green):
5´-antraquinone-TTT TTT TTT T-PC-TTT TTT TTT T-BMN Q535-3´(MW 7171 Da ).
The almost equally large decay products due to the PC linker placed in the middle of the sequence are highlighted in blue:
Product 1 5´-Phos-TTT TTT TTT T-BMN Q535-3´ (highlighted in light blue, MW 3536 Da) and product 2 5´-antraquinone-TTT TTT TTT T-PC modified-3´ (highlighted in dark blue, MW 3638 Da).
The decomposition products shown here were generated by a short laser pulse, the majority of the oligo remains uncleaved (green peak). The large left peak shows the double-protonated ion of the uncleaved conjugate (MW 3587 Da).
a) 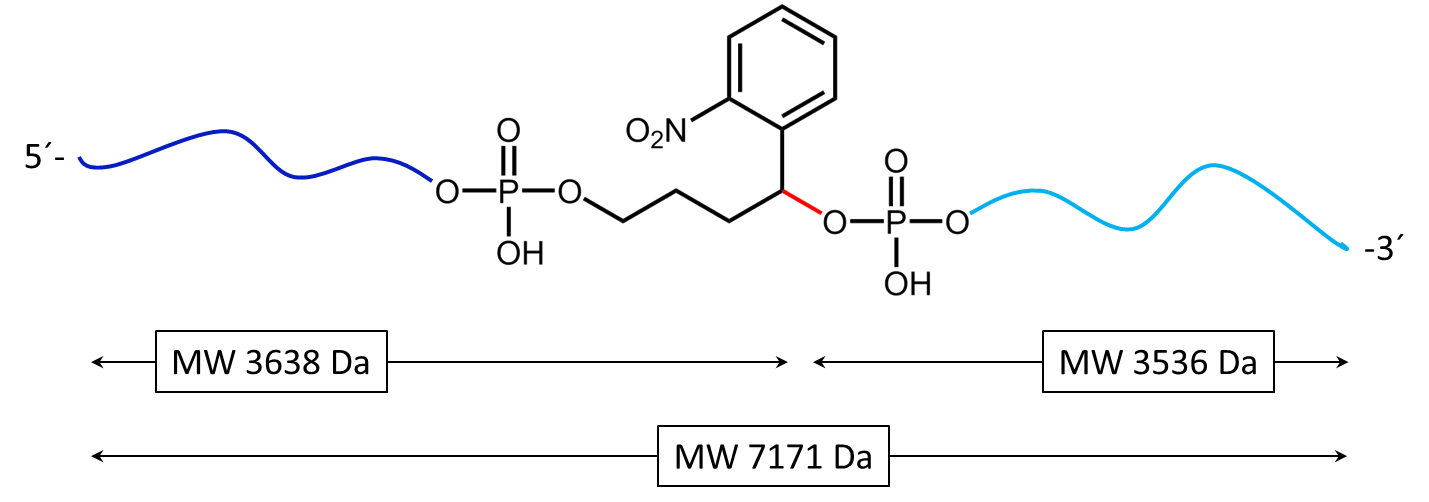
b) 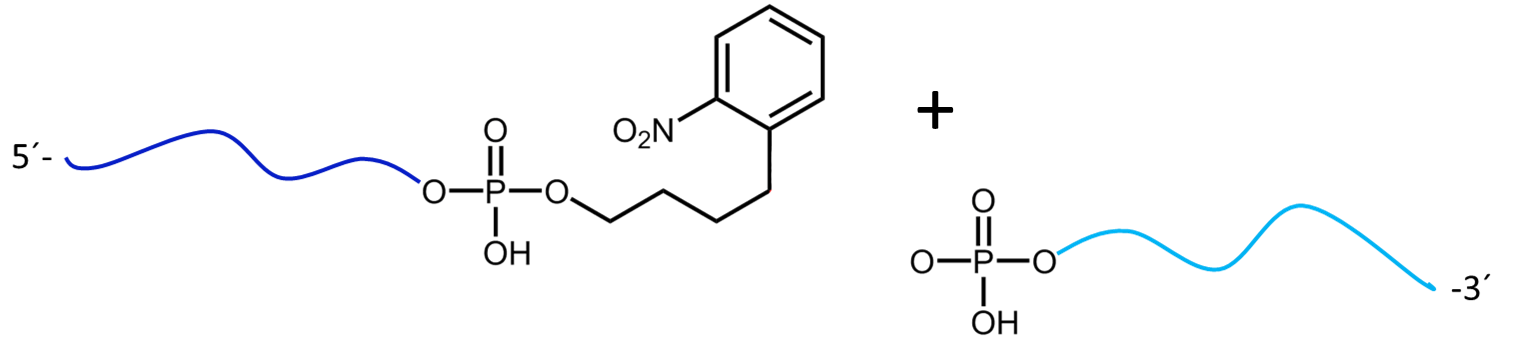
Figure 3a) shows the cleavage reaction of the oligo conjugate with Maldi laser irradiation. The cleavage site is shown in red. 3b) shows the resulting cleavage products after the partial decomposition of the PC linker.
For a complete cleavage of the photolabile linker in the oligo, light exposure takes longer, usually a few minutes. If the exposure is sufficiently long, the linker itself also disintegrates. In the following Figure 4, another cleavage product is shown after 30 minutes of exposure. Here, the linker which has already been cleaved completely falls off the oligo.
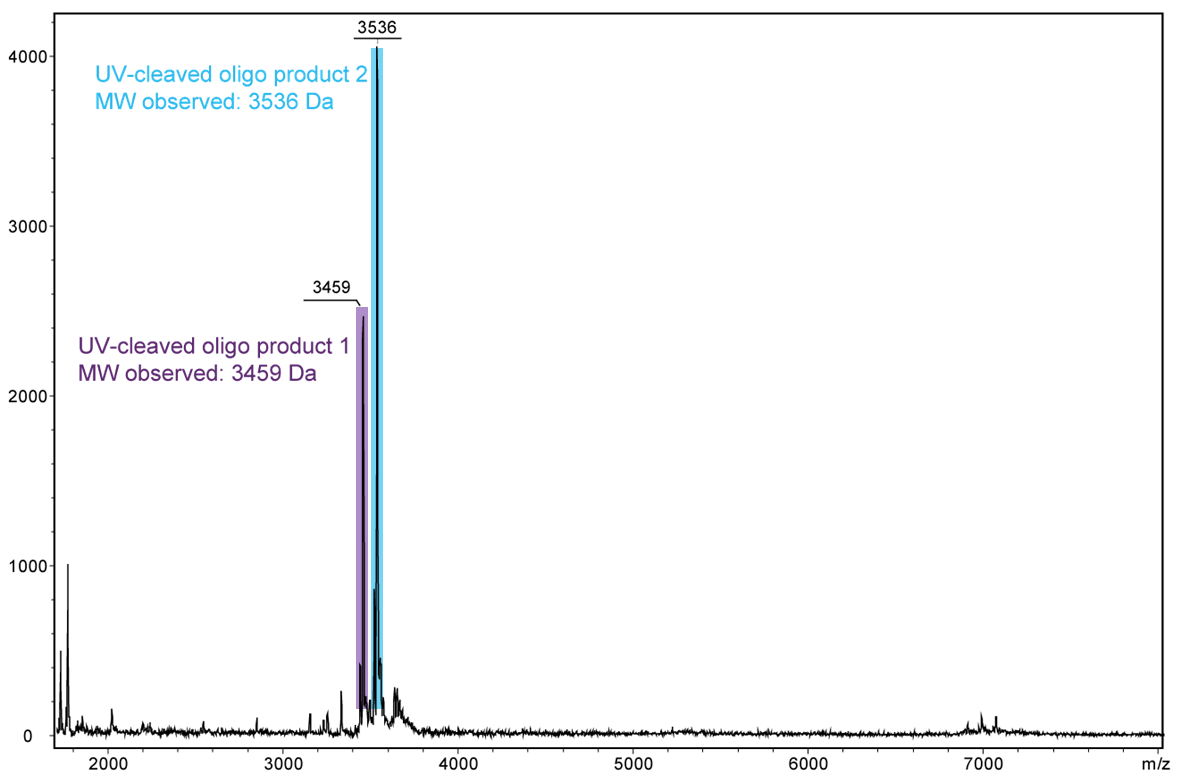
Figure 4) shows the result of the Maldi analysis of the oligo conjugate in aqueous solution after irradiation with UV light at 365 nm for 30 min. If the PC linker is irradiated with UV light, it disintegrates and completely falls off the oligo. The spectrum shows the two cleavage products of the conjugate: 5´-antraquinone-TTT TTT TTT T-Phos-3´ (highlighted in purple, 3459 Da) and 5´-Phos-TTT TTT TTT T-BMN Q535-3´ (highlighted in light blue, 3536 Da). The oligo conjugate is completely cleaved after UV exposure.
a) 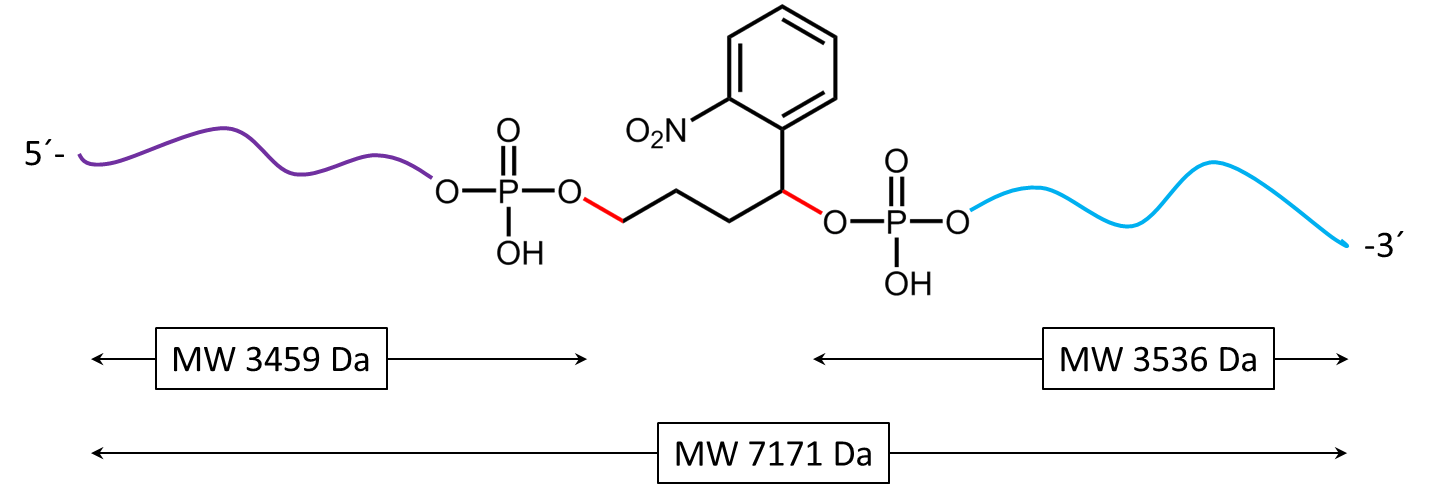
b)
Figure 5a) shows the cleavage reaction of the oligo conjugate when exposed to UV radiation. The cleavage sites are shown in red. 5b) shows the resulting cleavage products after the PC linker has completely disintegrated.
Literature:
1. Cleavable linkers in chemical biology. Leriche G, Chisholm L, Wagner A; Bioorg. Med. Chem. 20 (2012); 571–582.
2. Photocleavable linker for the patterning of bioactive molecules. Wegner SV, Sentürk OI, Spatz JP; Scientific Reports (2015), 5:18309.
3. Tag-Mass: Specific Molecular Imaging of Transcriptome and Proteome by Mass Spectrometry Based on Photocleavable Tag. Lemaire R, Stauber J, Wisztorski M, Van Camp C, Desmons A, Deschamps M, Proess G, Rudlof I, Woods AS, Day R, Salzet M, Fournier I; J Proteome Res. (2007), 6(6): 2057–2067.
4. Photocleavable peptide-DNA conjugates: synthesis and applications to DNA analysis using MALDI-MS. Olejnik J, Lüdemann HC, Krzymanska-Olejnik E, Berkenkamp S, Hillenkamp F, Rothschild KJ; Nucleic Acids Res. (1999), 27(23):4626-31.
5. Photocleavable biotin phosphoramidite for 5'-end-labeling, affinity purification and phosphorylation of synthetic oligonucleotides. Olejnik J, Krzymańska-Olejnik E, Rothschild KJ; Nucleic Acids Res. (1996), 24(2):361-6.
Photocrosslinker – labelling and cross linking of proteins and oligonucleotides
For effective cross-linking different photo-reactive groups are available that can be attached to the 5'-end of the oligonucleotide:
- Benzophenone C5
- Diazirine C5
- Psoralen
- Azidobenzoate C5
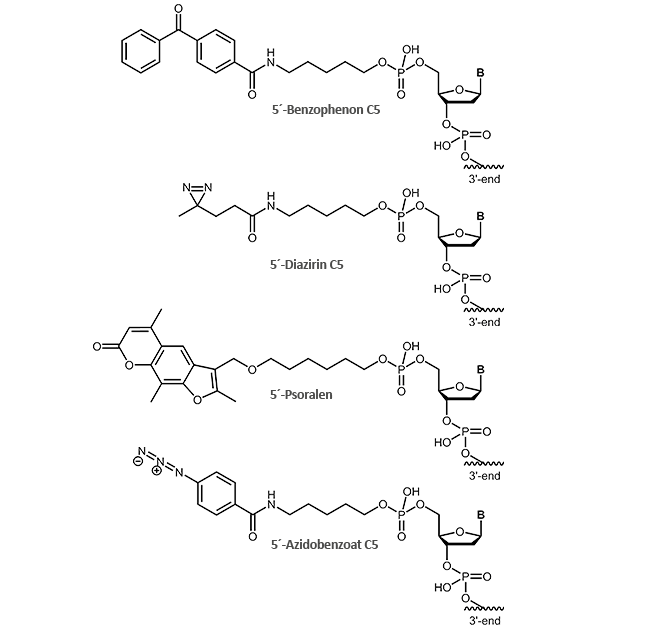
As the name suggests, photocrosslinkers are excited by light to produce a covalent bond between two molecules. The light induction is usually performed at wavelengths close to UV range (250-460 nm depending on the linker) and can be controlled precisely in time and space, e.g. only in certain tissue parts or at a defined developmental stage.
By binding the photocrosslinker to one end of an oligonucleotide, a complementary DNA section can be controlled with utmost precision. After photoinduction, a covalent bond is formed to the complementary strand. According to this, for example, a promoter region and the associated gene expression can be analysed, as well as the endogenous repair mechanisms which cells have developed in order to protect their DNA from UV damage.
As an alternative to conventional coupling strategies, photoreactive linkers also offer the possibility to bind functional groups to oligonucleotides.
Photocrosslinkers are also widely used to covalently link proteins and DNA.
DNA-protein-contacts are important switching points in cells, so accurate knowledge of the exact contact areas may allow modulation of such processes.
Literature:
1. Genetically encoded protein photocrosslinker with a transferable mass spectrometry-identifiable label. Yang Y, Song H, He D, Zhang S, Dai S, Lin S, Meng R, Wang C, Chen PR; Nat Commun. (2016), 7:12299. doi: 10.1038/ncomms12299.
2. Photolyase-like Repair of Psoralen-Crosslinked Nucleic Acids. Stafforst T, Hilvert D; Angew. Chem. Int. Ed. (2011), 50, 9483 –9486.
3. Sequence-specific photo-induced cross-linking of the two strands of double-helical DNA by a psoralen covalently linked to a triple helix-forming oligonucleotide. Takasugi M, Guendouz A, Chassignol M, Decout JL, Lhomme J, Thuong NT, Hélène C; Proc Natl Acad Sci U S A. (1991), 88(13):5602-6.
4. UV crosslinking of proteins to nucleic acids. Chodosh LA; Curr Protoc Mol Biol. (2001), Chapter 12:Unit 12.5. doi: 10.1002/0471142727.

