
DNA Oligonucleotides
Standard scales for oligonucleotides
Standard oligonucleotides up to 140 bases
Standard oligonucleotides are provided by biomers.net in four different scales. Here please find an overview of the offered length, yields, and purifications:
| Synthesis scale1 | XS 0,02 µmol |
S 0,05 µmol |
M 0,2 µmol |
L 1,0 µmol |
|---|---|---|---|---|
| Length (bases) | 12-60 | 12-140 | 6-140 | 6-140 |
| Wobbles | - | yes | yes | yes |
| Purification2 | ||||
| Cartridge | yes | yes | - | - |
| HPLC | - | yes | yes | yes |
| PAGE | - | yes | yes | yes |
| Yield3 | ||||
| Cartridge/HPLC | > 2 OD | > 3 OD | > 10 OD | > 30 OD |
| PAGE | - | > 1 OD | > 3 OD | > 10 OD |
| Quality control4 | ||||
| Maldi | yes | yes | yes | yes |
1. The synthesis scale defines the amount of starting material, it is not the yield of the finalised oligo. Due to the many steps of synthesis, processing and purification the final yield is significantly lower.
2. Oligos are delivery exclusively with purification. At least a cartridge purification is applied. We do not offer unpurified oligo nor only desalted oligos. By default all oligonucleotides with more than 60 bases in standard scales S - L are purified by HPLC (optional PAGE may be chosen).
3. The yields provided at "guaranteed delivery amount" are valid for a mixed 20mer DNA-sequence. For significantly shorter or longer oligos as well as for homopolymers this yield may differ. According to our experience, the actual delivery amounts exceed the guaranteed yields significantly, certainly we deliver the total yield.
4. Routinely, each oligo up to a length of 50 bases will be checked for quality by Maldi mass spectrometry.
Short to extra long oligos
2 - 5mer oligonucleotides
The actual synthesis process is identical for almost all lengths of oligonucleotides. Due to the small molecular size, very short oligos, however, differ in terms of the isolation and purification. We offer specific synthesis scales for di, tri, tetra and pentamer oligonucleotides including the corresponding adapted purification:
| Synthesis scale | DL2 | DL3 | DL4 | DL5 |
|---|---|---|---|---|
| Length (bases) | 2 | 3 | 4 | 5 |
| HPLC-purification | ja | ja | ja | ja |
| Maldi-quality control | ja | ja | ja | ja |
| Yield (µmol) approx. | 0,2 | 0,2 | 0,2 | 0,2 |
Oligonucleotides up to 200 bases
| Our new special synthesis CG scale includes: |
|---|
- Non-modified oligonucleotides between 140 and 200 bases
- Optimised synthesis conditions due to special cycles and chemicals
- Continuous monitoring of synthesis progress (trityl monitoring)
- Guaranteed yield > 250 pmol of purified oligonucleotide
- Inclusive 2-step purification: Reverse phase HPLC purification is followed by PAGE purification
How many correct clones can be expected ? We have analysed the results for you:
In cooperation with the University of Ulm, we exemplarily analysed a 200mer synthesised by biomers.net, which was amplified by PCR, cloned and finally sequenced. Out of 10 analysed clones 30% showed exactly the designated sequence. In a further clone a one base-insertion was found at position 22, in another the deletion of one base at position 147. In four additional clones several deletions were found. Due to synthesis errors, we expected mainly deletions of one or two bases, transitions and transversions as well as insertions. Interestingly, besides these expected changes, we found several deletions between 4 and 16 bases in a row. These can hardly be explained by synthesis errors, because the cyclic reaction of the synthesis is very unlikely to "skip" more than one or two bases. Likely the reason for these deletions is connected to the PCR reaction or the polymerase used (see also Hecker et al., 1998, BioTechniques 24:256-260).
In further analyses an average of 3 correct clones out of 5 has been found.
XXL synthesis DNA
Mid scale DNA synthesis (XXL)
for oligonucleotides amounts from 10 to 500 mg.
| Synthesis scale | XXL10MG | XXL25MG | XXL50MG | XXL100MG | XXL250MG | XXL500MG |
|---|---|---|---|---|---|---|
| Yield (mg) | > 10 mg | > 25 mg | > 50 mg | > 100 mg | > 250 mg | > 500 mg |
| Yield (OD), approx. | 400 OD | 850 OD | 1700 OD | 3400 OD | 8500 OD | 17000 OD |
| Yield (µmol) 20mer, approx. | 1,8 µmol | 3,5 µmol | 7,5 µmol | 15 µmol | 38 µmol | 77 µmol |
All XXL scale oligonucleotides are purified by reverse phase HPLC and quality checked by Maldi mass spectrometry.
The final amounts are approximate values and valid for a 20mer DNA oligonucleotide with a mixed sequence and not for homopolymers.
Wobbles
Wobbles
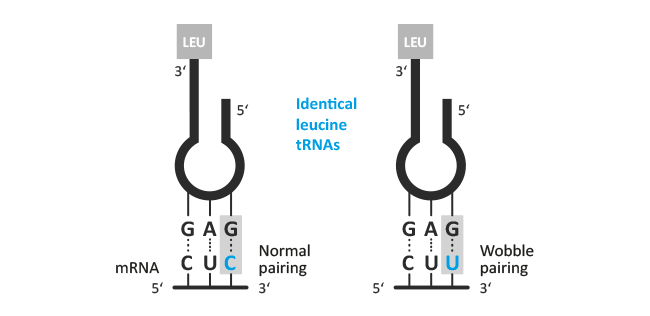
In molecular genetics, the wobble hypothesis explains how a particular tRNA can pair more than one mRNA triplet (codon). The first two positions of the codon usually bind to complementary bases of the anticodon of tRNA. Only the first position of the anticodon is not always specified and due to utilisation of rare bases like inosine, unusual base pairings are possible (wobble position).
Possible variants:
| Guanosine : Uridine | Inosine : Uridine | Inosine : Adenosine | Inosine : Cytidine |
 |
Oligonucleotides with wobbles (mixed bases, degenerate bases) can be used in many applications. During the synthesis of oligonucleotides wobbles can be incorporated at defined positions. Depending on the type of mixing base, specific statistical distributions of bases can be generated at each position in the oligo.
Synthesis of oligos with wobbles: Many modern DNA/RNA synthesizer “understand” the one letter code for wobbles (see below) and are able to mix the respective building blocks in the right proportion during the synthesis (2-wobble: 50% of each component, 3-wobble: 33% each and N=4-wobble: 25%). This quick and easy method usually leads to good results with small deviations from the desired statistical distribution. Depending on the particular sequence and the difference in reactivity of the building blocks, in individual cases the deviations can be higher. In critical cases or larger synthetic approaches manual mixing of the bases in the right proportion before the synthesis of the oligos can significantly improve the results. Furthermore, mixtures are also possible that deviate from a uniform distribution. For further information please contact our customer support team.
Purification of oligos with wobbles: Depending on the type and number of wobbles the synthesised oligonucleotide is composed of a variety of different sequences. These, possibly several thousand sequences may differ for example in their mobility in polyacrylamide gels (PAGE) or on the HPLC column. Oligos having many wobbles in their sequence lead to a broadening of the corresponding gel band or peak in the HPLC. Especially when cutting bands out of gel there is a risk that single sequences of the wobble mix get "lost". According to this we do not recommend PAGE purification of oligonucleotides with mixed bases, or only after consultation with our technical support team. The following internationally valid "one letter code" shows the possible wobble combinations of the four standard bases. Please use this code when ordering oligonucleotids with degenerate bases.
| Symbol | Explanation | Bases |
|---|
| Normal bases | |||||
| A | Adenine | A | |||
| C | Cytosine | C | |||
| G | Guanine | G | |||
| T | Thymine | T | |||
| U | Uracil (nur RNA) | U | |||
| Wobble bases | |||||
| W | Weak (binding bases) | A | T | ||
| S | Strong (binding bases) | C | G | ||
| M | aMino | A | C | ||
| K | Keto | G | T | ||
| R | puRine | A | G | ||
| Y | pYrimidine | C | T | ||
| B | Not A (B comes after A) | C | G | T | |
| D | Not C (D comes after C) | A | G | T | |
| H | Not G (H comes after G) | A | C | T | |
| V | Not T (V comes after T and U) | A | C | G | |
| N | aNy base | A | C | G | T |
Sequence modifications
Desoxyinosine (dI)
|
In addition to the four DNA bases, adenine, cytosine, guanine and thymine, the bases uracil and inosine and other rare nucleotides can be found in RNA. Inosine is an extremely rare nucleoside of RNA and it is composed of the sugar Despite the fact that inosine is a purine, inosine may enter into pairings with pyrimidines as well as purines of DNA. According to this, inosine is also known as "universal" or "neutral" base. However, some base pairings seem to be energetically more favorable than others, so that the strength of the bonds between the individual bases differs significantly. The strength of the bond decreases over the following series:1,2 |
 |
| The following table shows the possible mismatches of bases: |
|---|
| tRNA | A | C | G | U | I |
| mRNA | U | G | C | A | A |
| U | G | C | |||
| U | |||||
| G |
This wobble effect is also possible in oligonucleotides. Due to integration of an inosine nucleotide to the sequence of a primer, the primers obtain "variable" bases. These degenerate primers allow e.g. annealing and amplification of multiple related sequences. While using degenerate primers in PCR, DNA polymerases, such as Pfu, which have a so-called proof-reading function, have been reported to fall off the DNA as soon as they meet wobble bases in the sequence of the primer.3
Order information: Please be aware to enter deoxy-inosine, as well as ribo-inosine (2´-dInosine, rI) as internal modifications in the order. Identify the position of the internal modification in your sequence by a number (5,6,7,8), which mark the corresponding pull-down menu in the online order form. If no modifications are available for selection, neither the scale “XS” nor the purification “cartridge” must be selected for ribobases.
Literature: 1. Studies on the base pairing properties of deoxyinosine by solid phase hybridisation to oligonucleotides. Case-Green SC, Southern EM; Nucleic Acids Res. (1994), 22(2):131-6.
2. Base pairing involving deoxyinosine: implications for probe design. Martin FH, Castro MM, Aboul-ela F, Tinoco I Jr.; Nucleic Acids Res. (1985), 13: 8927-8938.
3. PCR with degenerate primers containing deoxyinosine fails with Pfu DNA Polymerase. Knittel T, Picard D, PCR Methods and Applications (1993), 2: 346-347.
Desoxyuridine (dU)
|
The nucleoside deoxyuridine (dU) consists of the sugar β-D-deoxyribose and the nucleobase uracil. Occasionally, deoxyuridine occurs in living cells by hydrolytic deamination of cytidine. The initiation of a repair program in the cell can prevent false pairing during replication of DNA. Order information: |
 |
More available sequence modifications can be found here .

