
Methylation
N6-methyl-adenosine
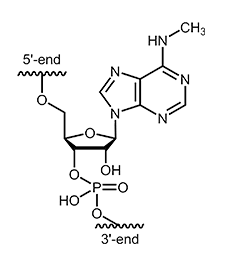
N6-methyl-adenosine (m6A)
|
N6-methyl-adenosine (m6A) is an abundant internal modification of natural mRNA strands in eukaryotes. Despite the discovery of m6A already in the 1970s, its biological function is still not completely clear and subject of current epigenetic research. |
 |
Literature:
1. N6-methyladenosine-dependent regulation of messenger RNA stability. Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, Ren B, Pan T, He C; Nature 505 (2014), 117–120
2. N6-methyl-adenosine (m6A) in RNA: An Old Modification with A Novel Epigenetic Function. Niu Y, Zhao X, Wu Y-S, Li M-M, Wang X-J, Yang Y-G; Genomics Proteomics Bioinformatics 11 (2013), 8–17.
3. Comprehensive Analysis of mRNA Methylation Reveals Enrichment in 3' UTRs and near Stop Codons. Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR; Cell 149 (2012), 1635–1646.
5-Me-dC and 5-hm-dC
5-Methyl-2'deoxycytidine and 5-Hydroxymethyl-2'deoxycytidine
biomers.net offers modified oligonucleotides with 5-Methyl-2'deoxycytidine (5-Me-dC)- as well as 5-Hydroxymethyl-2'deoxycytidine (5-hm-dC).
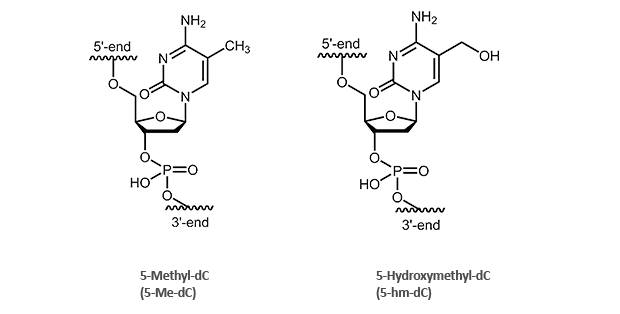 |
5-Methyl-2’-desoxycytidine (5-Me-dC)
Methylation on cytosine bases is a common mechanism in eukaryotic cells for the regulation of chromosomal activity; the most prominent example in this regard is surely the "silencing" of whole chromosoms.
The research area "Epigenetic" studies such mechanisms by which genetic information is regulated and controlled beyond the plain sequence.
The methylation state of the DNA is carefully controlled by the cells and has important influence on their activity and behaviour.
Particularly in tumor research the methylation state of genome has a prominent position, as findings indicated differences in the methylation pattern of tumor cells and normal cells. This different pattern accounts for different genetic activities, which play a crucial role in tumor development and progression.
To analyse the exact regulative processes, often the exact position of the methylation is required, best directly accessible. This may be achieved with oligonucleotides bearing a methylated cytosine in the required position.
5-Hydroxymethyl-2’-desoxycytidine (5-hm-dC)
Already in 1952 Wyatt and Cohen succeeded in isolating 5-hydroxymethylcytosin from bacteriophage DNA1. However, recently this socalled "sixth base" moved into the focus of epigenetic research when significant amounts of 5-hm-dC were found in different cell tissue DNA (brain, liver)2,3.
Apparently it is not possible to differentiate between 5-Me-dC and 5-hm-dC by the usual "bisulfite sequencing" which is frequently used for methylation studies4. Accordingly there is a need for new technologies to (re)evaluate the grade of DNA methylation in detail. So, for example, 5-hm-dC can be selectively glycosylated by enzymatic reactions5,6, or oxidized under mild conditions as recently described by Booth et al.7. Both procedures allow to distinguish between 5-hm-dC and 5-Me-dC in further analytical analysis steps.
Literature:
1. A new pyrimidine base from bacteriophage nucleic acids. Wyatt GR, Cohen SS; Nature (1952), 170: 1072.
2. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Kriaucionis S, Heintz N; Science (2009), 324: 929.
3. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Roa A; Science (2009), 324: 930.
4. Enzymatic approaches and bisulfite sequencing cannot distinguish between 5-methylcytosine and 5-hydroxymethylcytosine in DNA. Nester C, Ruzov A, Meehan RR, Dunican DD, BioTechniques (2010), 48, 317.
5. A novel method for the efficient and selective identification of 5-hydroxymethylcytosine in genomic DNA. Robertson AB, Dahl JA, Vagbo CB, Tripathi P, Krokan HE, Klungland A, Nucleic Acids Research (2011), 39, e55.
6. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Song CX, Szulwach KE, Fu Y, Dai Q, Yi C, Li X, Li Y, Chen C, Zhang W, Jian X, Wang J, Zhang L, Looney TJ, Zhang B, Godley LA, Hicks LM, Lahn BT, Jin P, He C, Nature Biotechnology (2011), 29, 68.
7. Quantitative Sequencing of 5-Methylcytosine and 5-Hydroxymethylcytosine at Single-Base Resolution. Booth MJ, Branco MR, Ficz G, Oxley D, Krueger F, Reik W, Balasubramanian S; Science (2012), 336, 934.

